 |
 |
| Anim Biosci > Volume 32(10); 2019 > Article |
|
Abstract
Objective
Methods
Results
Conclusion
ACKNOWLEDGMENTS
Figure 1
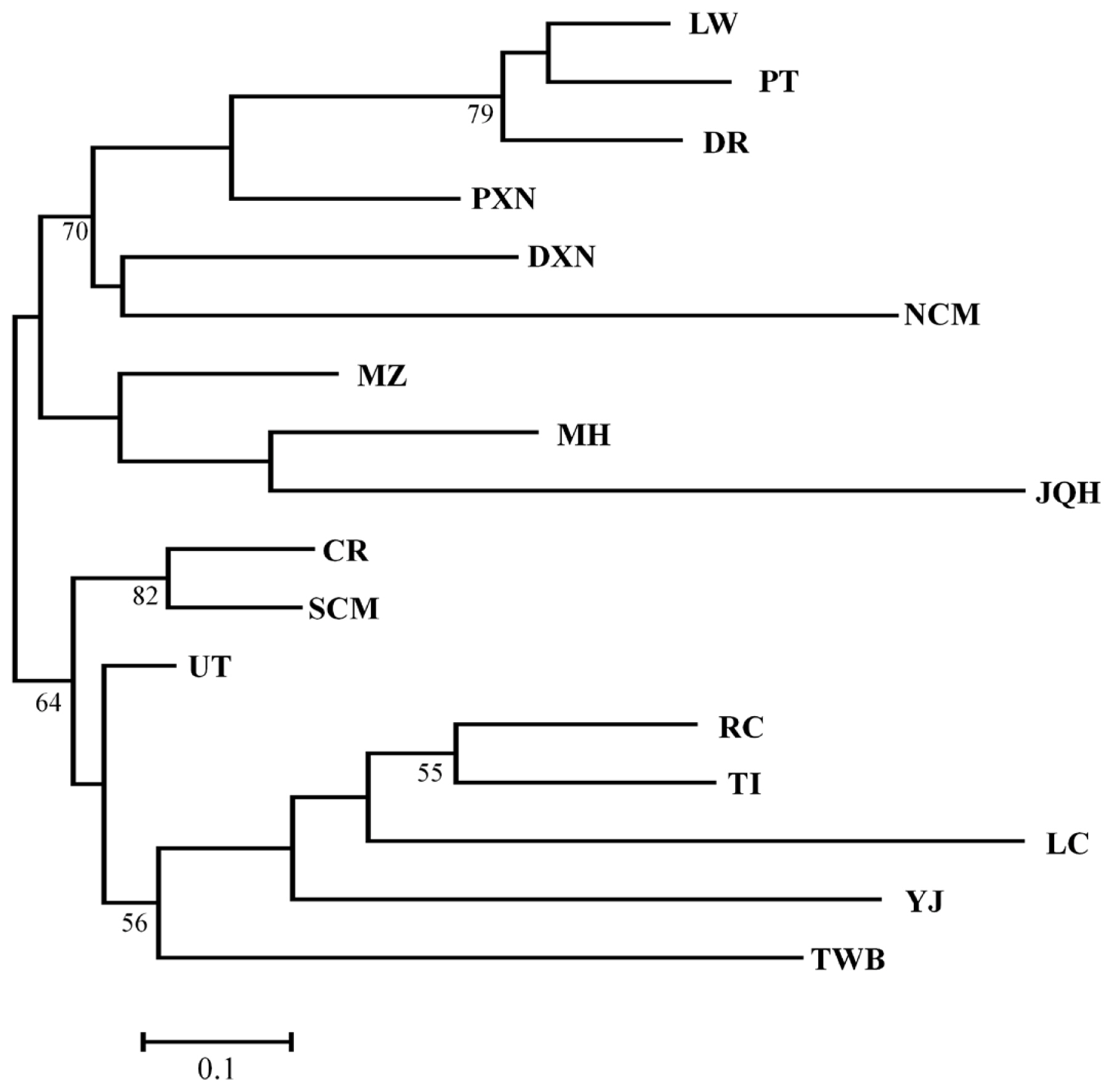
Figure 2
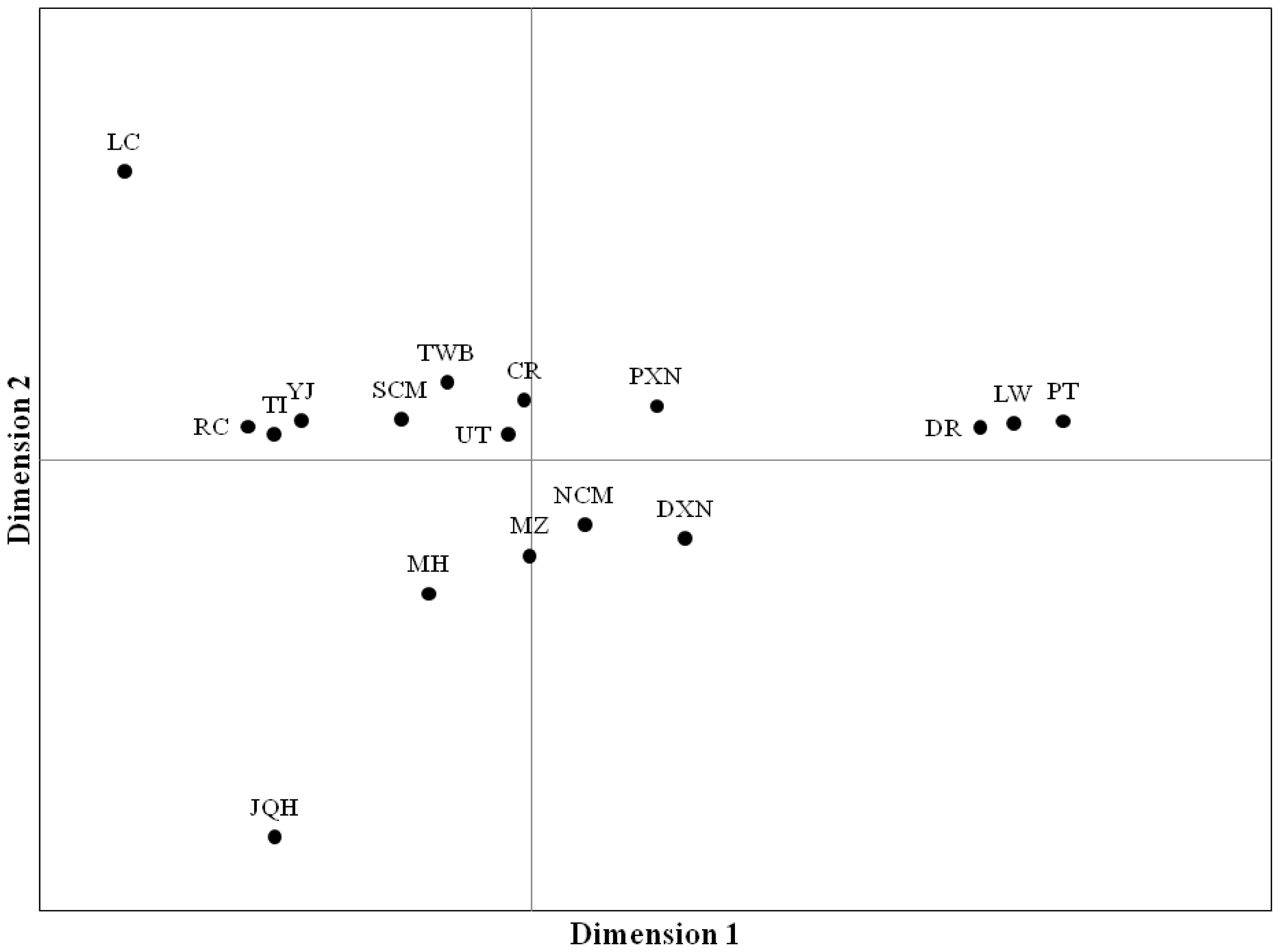
Figure 3

Table 1
Table 2
| Locus | SSC | TNA | MNA | Ne | PIC | Ho | He | FIT | FST | FIS | HWE | Private allele; bp (animal and frequency) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S0155 | 1 | 10 | 5.73 | 3.58 | 0.835 | 0.736 | 0.733 | 0.139 | 0.179 | −0.049 | NS | 143 (UT 0.05) |
| CGA | 1 | 29 | 10.64 | 7.15 | 0.914 | 0.761 | 0.889 | 0.171 | 0.072 | 0.107 | ** | |
| S0226 | 2 | 11 | 4.45 | 3.04 | 0.761 | 0.785 | 0.669 | 0.013 | 0.193 | −0.224 | * | |
| SW240 | 2 | 14 | 5.36 | 3.60 | 0.828 | 0.741 | 0.708 | 0.127 | 0.199 | −0.091 | ** | 120 (UT 0.07) |
| S0002 | 3 | 27 | 7.91 | 5.25 | 0.896 | 0.839 | 0.794 | 0.068 | 0.155 | −0.103 | NS | 204 (UT 0.05); 252 (NCM 0.05) |
| SW72 | 3 | 10 | 5.36 | 2.95 | 0.697 | 0.617 | 0.656 | 0.141 | 0.124 | 0.020 | NS | |
| S0227 | 4 | 9 | 4.09 | 2.85 | 0.707 | 0.683 | 0.648 | 0.068 | 0.154 | −0.102 | NS | 229 (TWB 0.14) |
| IGF-1 | 5 | 10 | 5.36 | 3.84 | 0.795 | 0.765 | 0.738 | 0.060 | 0.131 | −0.082 | NS | |
| S0005 | 5 | 19 | 7.00 | 4.73 | 0.861 | 0.671 | 0.810 | 0.241 | 0.123 | 0.134 | ND | |
| SW122 | 6 | 13 | 5.64 | 3.49 | 0.845 | 0.728 | 0.712 | 0.156 | 0.208 | −0.066 | NS | 99 (TWB 0.09); 107 (TWB 0.23); 109 (TWB 0.09) |
| S0101 | 7 | 11 | 4.73 | 2.89 | 0.778 | 0.633 | 0.654 | 0.234 | 0.241 | −0.009 | ** | |
| SW632 | 7 | 11 | 6.00 | 3.58 | 0.823 | 0.635 | 0.732 | 0.248 | 0.168 | 0.095 | ** | |
| S0225 | 8 | 13 | 5.09 | 3.34 | 0.767 | 0.778 | 0.700 | 0.017 | 0.152 | −0.160 | NS | 173 (NCM 0.05) |
| SW911 | 9 | 10 | 6.00 | 3.87 | 0.818 | 0.734 | 0.729 | 0.112 | 0.152 | −0.047 | * | 144 (CR 0.09) |
| SW951 | 10 | 7 | 3.82 | 2.69 | 0.651 | 0.554 | 0.645 | 0.210 | 0.119 | 0.103 | * | |
| S0386 | 11 | 9 | 4.18 | 2.84 | 0.663 | 0.293 | 0.635 | 0.583 | 0.135 | 0.518 | ** | |
| S0090 | 12 | 10 | 5.18 | 3.48 | 0.814 | 0.785 | 0.701 | 0.062 | 0.196 | −0.167 | NS | |
| S0068 | 13 | 17 | 7.45 | 5.08 | 0.884 | 0.878 | 0.802 | 0.018 | 0.141 | −0.142 | NS | 255 (SCM 0.16) |
| S0215 | 13 | 13 | 4.45 | 3.07 | 0.737 | 0.553 | 0.580 | 0.251 | 0.246 | 0.007 | ** | |
| SW857 | 14 | 11 | 5.73 | 4.11 | 0.852 | 0.633 | 0.755 | 0.269 | 0.163 | 0.127 | NS | |
| S0355 | 15 | 12 | 5.82 | 3.80 | 0.815 | 0.685 | 0.697 | 0.179 | 0.199 | −0.025 | NS | |
| SW936 | 15 | 18 | 6.09 | 4.12 | 0.836 | 0.840 | 0.764 | 0.029 | 0.153 | −0.146 | ** | |
| S0026 | 16 | 9 | 5.09 | 3.35 | 0.762 | 0.731 | 0.715 | 0.083 | 0.140 | −0.067 | NS | |
| SW1031 | 17 | 11 | 4.82 | 2.99 | 0.696 | 0.498 | 0.659 | 0.319 | 0.137 | 0.211 | ** | 116 (DR 0.07) |
| S0120 | 18 | 15 | 7.00 | 4.34 | 0.797 | 0.773 | 0.784 | 0.062 | 0.089 | −0.029 | NS | |
| S0218 | X | 11 | 4.09 | 2.62 | 0.685 | 0.335 | 0.562 | 0.532 | 0.249 | 0.377 | ** | |
| Mean | 14.59 | 5.65 | 3.71 | 0.789 | 0.679 | 0.710 | 0.169 | 0.162 | 0.007 |
SSC, Sus scrofa chromosome; TNA, total number of alleles per locus; MNA, mean number of alleles per locus; Ne, effective number of alleles per locus; PIC, polymorphism information content, Ho and He, the observed and unbiased expected heterozygosity; FIT, inbreeding coefficient of an individual relative to the total population; FST, effect of subpopulations compared with the total populations; FIS, inbreeding coefficient of an individual relative to the subpopulation; HWE, Hardy-Weinberg equilibrium; NS, no significant difference; ND, not done.
Table 3
Table 4
MH, Mae Hongson; SCM, Southern part of Chiang Mai; NCM, Northern part of Chiang Mai; CR, Chiang Rai; UT, Uttaradit; TWB, Thai wild boars; DR, Duroc; LW, Large White; PT, Pietrain; DXN, Duroc×native crossbred pig; PXN, Pietrain×native crossbred pig; JQH, Jiangquhai; LC, Luchuan; MZ, Min; YJ, Yushanhei; TI, Tibetan; RC, Rongshang.
REFERENCES
- TOOLS
-
METRICS

- Related articles
-
Genetic diversity and population genetic structure of Cambodian indigenous chickens2022 June;35(6)






 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print




