 |
 |
Abstract
ACKNOWLEDGEMENTS
Figure 1

Figure 2
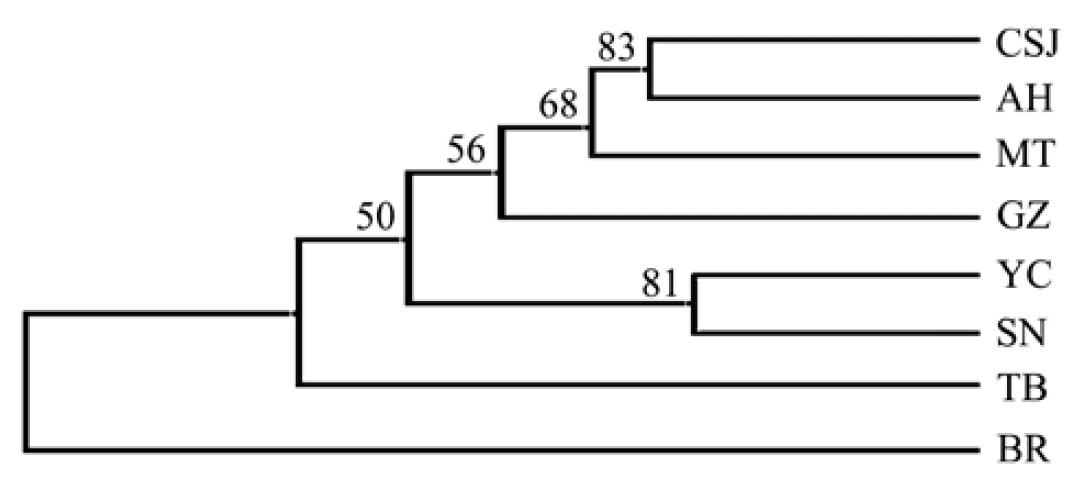
Figure 3

Table 1
Table 2
Tm = Annealing temperature; At = Total number of alleles per locus; Rt = Allelic richness over all samples.
Hoffmann, I., P. A. Marsan, S. F. Barker, E. G. Cothran, O. Hanotte, J. A. Lenstra, D. Milan, S. Weigend and H. Simianer. 2004. New MoDAD marker sets to be used in diversity studies for the major farm animal species: recommendations of a joint ISAG/FAO working group. In: 29th International Conference on Animal Genetics. FAO, Meiji University, Tokyo, Japan.
Table 3
| Breeds and codes | Allelic diversity | Genetic diversity | FIS | |||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| TNA | MNA | NEA | NPA | AR | Ĥ (SD) | Ho(SD) | ||
| Tibet goat (TB) | 69 | 6.27 | 3.93 | 2 | 6.21 | 0.738(0.013) | 0.697(0.022) | −0.056* |
| Guizhou white goat (GZ) | 75 | 6.82 | 4.25 | 3 | 6.82 | 0.764(0.010) | 0.711(0.020) | 0.071** |
| Shannan white goat (SN) | 77 | 7.00 | 4.32 | 4 | 6.88 | 0.759(0.016) | 0.706(0.021) | 0.069** |
| Yichang white goat (YC) | 73 | 6.64 | 4.20 | 8 | 6.59 | 0.748(0.011) | 0.711(0.016) | −0.051** |
| Matou goat (MT) | 77 | 7.00 | 4.55 | 4 | 6.94 | 0.781(0.014) | 0.738(0.020) | 0.036 |
| Changjiangsanjiaozhou white goat (CSJ) | 74 | 6.73 | 4.49 | 2 | 6.64 | 0.774(0.014) | 0.723(0.016) | 0.066*** |
| Anhui white goat (AH) | 81 | 7.36 | 4.59 | 5 | 7.35 | 0.771(0.011) | 0.718(0.021) | 0.063 |
| Mean | 6.78 | 0.762(0.014) | 0.715(0.017) | |||||
| Boer goat (BR) | 67 | 6.09 | 4.15 | 3 | 6.07 | 0.734(0.013) | 0.634(0.024) | −0.137*** |
| ALL Mean | 6.69 | 0.759(0.013) | 0.705(0.018) | |||||
TNA = Total number of alleles; NEA = Number of effective alleles; MNA = Mean number of alleles; AR = Allelic richness; NPA = Number of private alleles; Ĥ = Unbiased gene diversity; Ho = Observed heterozygosity; SD =Standard deviation; HWE = Number of loci that deviate from a Hardy Weinberg equilibrium; FIS = Population inbreeding coefficient.
REFERENCES
- TOOLS
-
METRICS

- Related articles
-
Genetic diversity of Indonesian cattle breeds based on microsatellite markers2019 April;32(4)





 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print




