 |
 |
| Anim Biosci > Volume 37(2); 2024 Special Issue > Article |
|
Abstract
Rumen microbiota play a central role in the digestive process of ruminants. Their remarkable ability to break down complex plant fibers and proteins, converting them into essential organic compounds that provide animals with energy and nutrition. Research on rumen microbiota not only contributes to improving animal production performance and enhancing feed utilization efficiency but also holds the potential to reduce methane emissions and environmental impact. Nevertheless, studies on rumen microbiota face numerous challenges, including complexity, difficulties in cultivation, and obstacles in functional analysis. This review provides an overview of microbial species involved in the degradation of macromolecules, the fermentation processes, and methane production in the rumen, all based on cultivation methods. Additionally, the review introduces the applications, advantages, and limitations of emerging omics technologies such as metagenomics, metatranscriptomics, metaproteomics, and metabolomics, in investigating the functionality of rumen microbiota. Finally, the article offers a forward-looking perspective on the new horizons and technologies in the field of rumen microbiota functional research. These emerging technologies, with continuous refinement and mutual complementation, have deepened our understanding of rumen microbiota functionality, thereby enabling effective manipulation of the rumen microbial community.
The rumen microbiome is indispensable for the survival, productivity, and overall health of ruminant animals, as the microbiome plays a vital role in their early development, health, and physiological processes. The diversity of rumen microorganisms results from mutual selection and coevolution between the microorganisms and their hosts, resulting in a dynamic balance of interdependence and restriction [1]. On the one hand, the host provides a favorable living environment and fermented substrates for the growth of rumen microorganisms [2]. On the other hand, rumen microbes play a crucial role in the breakdown of plant cellulose, hemicellulose, starch, and other components, providing energy and essential nutrients to the host for survival and production. Therefore, investigating the types and functions of rumen microorganisms, as well as the interactions between these microorganisms and their host, holds great significance in enhancing the production performance of ruminants.
For a long time, our knowledge on the rumen microbiome has relied primarily on iso lation and culture-based methods. Although this approach is limited when culturing the entire spectrum of rumen microbes, it can provide valuable insights into the accurate functional capacities of specific microbial members (Table 1). In recent years, however, researchers have focused more on omics-based approaches in rumen microbiology research (Figure 1) [3ŌĆō5]. Through this method, microbial community phylogeny, diversity, composition, and functional capabilities can be explored in a culture-independent and high-throughput manner, which has significantly expanded our knowledge on the rumen microbiome and its ecological roles.
Studies have shown that while the composition of rumen microbial communities may vary across studies, the metabolic and functional aspects of these communities remain relatively stable [6]. This reveals a significant inconsistency between the current knowledge on rumen microbial taxonomy and functional annotations. There are two potential reasons for this discrepancy. First, there is a deviation between the principles of species taxonomy and functional classification. For example, two strains from the same species may perform distinct metabolic functions, while different functions may be shared by multiple microbial lineages [7]. Second, when translating species taxonomy annotations into metabolic functions, the redundancy and variability of these functions become evident [8]. Therefore, it is essential to recognize the limitations of focusing solely on species taxonomy [9]. Instead, attention should be directed toward the functional repertoire of the rumen microbiota to reveal the true events.
Among all livestock species, ruminant animals are the most efficient in utilizing fiber due to their unique digestive system, the rumen microbiota. The cellulolytic bacterium Fibrobacter succinogenes was initially identified in the rumen of cows in 1947 [10]. Moreover, by assessing the potential for cellulose degradation through enzyme catalysis in pure isolates and employing relative quantification real-time polymerase chain reaction (PCR), F. succinogenes was revealed as the predominant bacterium in terms of efficiency and prevalence in cellulose degradation within the rumen [11]. Another cellulolytic bacterium, Ruminococcus flavefaciens, exhibits comparable abundance to F. succinogenes in lactating cows under conventional feed conditions [12]. Under cellulose-restricted conditions, R. flavefaciens becomes dominant. The third most common group of cellulolytic bacteria is R. albus. Compared to F. succinogenes and R. flavefaciens, R. albus is less abundant, but it is particularly competitive in the initial adhesion to cellulose and growth [13]. Other cellulolytic bacteria have also been cultured and well studied. For example, Clostridium lochheadii [14] is highly active in digesting cellulose. C. longisporum, Eubacterium cellulosolvens, and Butyrivibrio fibrisolvens [15] are also cellulolytic bacteria isolated from the rumen.
In addition to bacteria, certain protozoa exhibit cellulolytic activity, which may arise from ingested fibrolytic microorganisms or their own fiber-degrading enzymes. Among the rumen protozoa studied, Eudiplodinium maggii, Epidinium ecaudatum, and Ostracodinium dilobum emerged as the most efficient cellulose degraders [11]. Nevertheless, exploring this capability is challenging due to the complexities associated with maintaining ruminal protozoa in culture [16]. Anaerobic fungi have also been successfully isolated from various ruminant species, showcasing their impressive capacity for cellulose hydrolysis [17]. Despite their relatively low numerical abundance, these anaerobic fungi play a significant role in cellulose degradation within the rumen [18].
Compared to cellulose, the structure of hemicellulose is more complex and easier to hydrolyze; thus, research on hemicellulose degradation is limited. All cellulolytic bacteria can degrade hemicellulose. In early experiments, in addition to cellulolytic bacteria, Lachnospira multiparus, and Prevotella ruminicola isolated and cultured from the rumen could degrade hemicellulose [19]. Prevotella species, although less efficient than B. fibrisolvens [20], play a crucial role in ruminal hemicellulose degradation due to their prevalence as very common microbes in the rumen. Notably, the degradation capabilities of different hemicellulolytic bacteria are specialized due to the different structures of hemicellulose. For example, B. fibrisolvens is more effective in degrading xylan than other hemicelluloses (xyloglucan, glucomannan, and ╬▓-glucan) [20]. The degradation of glucomannan is primarily carried out by Streptococcus species [20]. Three ruminal protozoa species (Polyplastron multivesiculatum, Eudiplodinium maggii, and Entodinium species) can also degrade hemicellulose due to their endo-xyloglucanase and endo-xylanase activities [21]. However, most of the discovery of fungi is based on metagenomics, which should be researched more through culture.
The rumen has a large population of amylolytic bacteria, such as S. bovis, P. ruminicola, Ruminobacter amylophilus, Succinimonas amylolytica, and Selenomonas ruminantium [22,23]. Notably, resistant starch is degraded by the specialized R. bromii [24]. Some cellulolytic bacteria can also utilize starch, including B. fibrisolvens, F. succinogenes, and Clostridium species [25,26]. Amylolytic bacteria breakdown starch into oligosaccharides, which are usually directly fermented into volatile fatty acids (VFAs). However, for some amylolytic bacteria, the fermentation end products are intermediate molecules. For example, S. bovis primarily produces lactate as the end product [23], while R. amylophilus produces succinate during amylolysis [22]. These lactate and succinate molecules subsequently undergo further fermentation by other bacteria into VFAs. Approximately 20% to 45% of starch degradation activity in the rumen is attributed to protozoa [27]. Protozoal species including Eremoplastron bovis, Diploplastron affine, Ophryoscolex caudatus, and Polyplastron multivesiculatum exhibit a remarkable capacity for starch degradation [28]. Additionally, rumen fungi like Orpinomyces joyonii, Neocallimastix patriciarum, and Piromyces communis can digest cereal starches [29]. However, the presence of protozoa and fungi for starch degradation in the rumen is not essential.
Pectin can be fully digested by bacteria and protozoa, and its degradation rate in the rumen is higher than that of other carbohydrates. Pectin-degrading bacteria include B. fibrisolvens, P. ruminicola, L. multipara, S. bovis, Succinivibrio dextrinosolvens, and Treponema saccharophilum [30]. Additionally, the common cellulolytic bacteria R. albus and F. succinogenes can degrade pectin [31]. Among them, the primary pectin-degrading bacteria are B. fibrisolvens and Prevotella species [32]. Notably, similar to ciliate protozoa, S. bovis can degrade pectin but cannot utilize the degradation products.
This protein degradation process involves the participation of many bacteria, and proteolytic and peptidolytic bacteria account for approximately 65% of rumen bacteria [33]. The most important proteolytic bacteria are B. fibrisolvens and B. proteoclasticus [34], which exhibit high proteolytic capacity, and the abundance of B. fibrisolvens increases when animals are fed high-protein diets [35]. Additionally, R. amylophilus, E. budayi, Streptococcus bovis, Selenomonas ruminantium [36], and other bacteria also exhibit proteolytic capabilities. While these bacteria may be present at lower abundances in the rumen, they play a crucial role in protein metabolism due to their high proteolytic abilities. Prevotella species, such as P. albensis, P. brevis, and P. bryantii, also possess proteolytic capabilities [37]. Although their proteolytic capacity may be lower than that of other bacteria, these species are important contributors to protein degradation due to their higher abundance in the rumen. Protozoa contribute to approximately 20% of the proteolytic activity in the rumen [38]. Protozoa such as Entodinium caudatum, Entodinium simplex, Dasytricha ruminantium, and Polyplastron multivesiculatum, exhibit proteolytic abilities [39]. Studies have isolated the fungus Neocallimastix frontalis from sheep rumen, which exhibits high proteolytic activity and plays an important role in rumen protein degradation [40]. However, other studies comparing the activities of rumen proteolytic fungi suggest that the capacity of these fungi for protein hydrolysis is limited.
Peptide degradation is an intermediate step in protein breakdown, and Prevotella species exhibit broad peptidolytic activity. Among these, P. ruminicola stands out as a pivotal species for peptide degradation in the rumen, exhibiting a dipeptidyl peptidase range and specific activity that surpasses those of other prevalent peptidolytic bacteria [37]. Additionally, P. albensis and P. bryantii possess peptidase activity. Apart from Prevotella species, S. bovis, R. amylophilus, Veillonella parvula, Ruminococcus species, Megasphaera elsdenii, L. multipara, F. succinogenes, and E. ruminantium [41] exhibit weaker peptidase activity, contributing less to peptide degradation in the rumen. In the absence of bacteria, ciliate plays a significant role in the accumulation and breakdown of dipeptides [42], for example, Entodinium species, Dasytricha ruminantium, and Isotricha species can degrade oligopeptides in the rumen.
Lipids in the rumen are hydrolyzed into galactose, glycerol, and long-chain or medium-chain fatty acids. The first identified lipid-degrading bacterium in the rumen was Anaerovibrio lipolyticus [43], followed by the isolation of various other lipid-degrading bacteria, including B. fibrisolvens, Clostridium species, and Propionibacterium species [44]. The prominent lipid-degrading bacteria include A. lipolytica and B. fibrisolvens. Different lipid-degrading bacteria exhibit varying abilities and preferences for specific types of lipids. For instance, B. fibrisolvens can only degrade polar lipids, while Propionibacterium species can only degrade neutral lipids [44]. There is limited research on fungi and protozoa involved in lipid degradation. Some studies have found that the protozoan species Entodinium caudatum has phospholipase activity, but its relevance to dietary lipids remains uncertain [45]. Reports suggest that the protozoa Epidinium species account for 30% to 40% of the lipid degradation activity in the rumen, but it is generally believed that bacteria are the primary species responsible for lipid degradation [46]. In the rumen, the breakdown of unsaturated fatty acids involves their hydrogenation into saturated fatty acids. This process may serve as a detoxification mechanism because unsaturated fatty acids are more toxic to microorganisms than saturated fatty acids [47]. Bacteria and protozoa participate in the process of biohydrogenation, and these bacteria include B. hungatei, B. proteoclasticus, Propionibacterium acnes, E. ruminantium, C. proteoclasticum, Pseudobutyrivibrio species [44,48]. These bacteria can be divided into the following groups: one can convert linoleic acid to trans-11-octadecenoic acid, and the other produces stearic acid as the end product.
Rumen microorganisms primarily ferment soluble sugars, amino acids, and glycerol to generate products such as VFAs. The majority of the rumen microbiome, including the genera Streptococcus, Bifidobacterium, Lactobacillus, Treponema, Selenomonas, Veillonella, Coprococcus and Megasphaera [6, 49], can ferment these soluble sugars. Among them, S. bovis and Lactobacillus species are important, as they can rapidly proliferate in the presence of excess carbohydrates [48]. Succinate, lactate, and fumarate are intermediate products in the fermentation process and eventually convert into VFAs. One of the pathways is called the succinate pathway, which involves the reduction of pyruvate to produce succinate and its subsequent conversion to propionate. Bacteria from the phyla Firmicutes and Bacteroidetes are involved in the succinate pathway [6]. Specifically, bacteria such as Actinobacillus succinogenes and Mannheimia succiniciproducens are responsible for succinate production in the rumen [50], while Succiniclasticum ruminis is among the succinate-utilizing bacteria [51]. Another pathway, the acrylate pathway, converts lactate into propionate, among others. This pathway is vital in the rumen, as it prevents the accumulation of lactate, which can lead to acidosis due to a decrease in ruminal pH. Common lactate-utilizing bacteria include S. ruminantium and M. elsdenii, which ferment lactate to acetate and propionate [52]. Bacteria from the genera Lactobacillus, Streptococcus, Enterococcus, and Pediococcus are primarily responsible for lactate production.
Most of the amino acids are rapidly fermented in the ru men; the first step produces ammonia and keto acids, which are then converted into VFAs. Almost all proteolytic bacteria are involved in the deamination process. Deaminating bacteria can be divided into two main categories. The first category includes bacteria with lower deamination capability but high abundance in the rumen [53], such as B. fibrisolvens, P. ruminicola, M. elsdenii, and Allisonella histaminiformans. The second category consists of bacteria with strong deamination capability but low abundance, also known as high-ammonia-producing bacteria. These bacteria include C. aminophilum, C. sticklandii, and Peptostreptococcus anaerobius. Notably, high-ammonia-producing bacteria cannot participate in protein degradation and can only ferment amino acids as their only nitrogen source. Glycerol broken down from lipids is rapidly fermented in the rumen into VFAs, CO2, and H2. A. lipolytica, B. fibrisolvens, and S. ruminantium are involved in this fermentation process [48].
The hydrolysis and fermentation processes of macromolecules generate a large amount of hydrogen, which can be converted into methane in the rumen. Methanogens are considered a key driving force in the entire food chain [54]. Methanogens that have been cultured from rumen contents include Methanobacterium formicium, Methanobacterium bryantii, Methanobrevibacter olleyae, Methanobrevibacter millerae, Methanobrevibacter ruminantium, Methanomicrobium mobile, Methanoculleues olentangyi, and Methanosarcina barkeri [48]. Among them, the most prevalent methanogen genus was Methanobrevibacter, constituting 66% to 68% of the archaeal population [55]. The most distinctive methanogens are Methanobrevibacter ruminantium [48], and Methanobrevibacter gottschalkii, and Methanobrevibacter ruminantium are generally dominant.
Methanogenic archaea can be classified into the following types based on different substrates: H2/CO2 (hydrogenotrophic), methane derivatives (methylotrophic), and acetate (acetoclastic). The hydrogenotrophic pathway is the main route for methane production, with approximately 78% of methanogens participating in this process [49]. The most important hydrogenotrophic methanogen genera are Methanobrevibacter, Methanosphaera, Methanimicrococcus, and Methanobacterium [56]. The methylotrophic pathway involves the simultaneous utilization of methyl compounds, for growth. Approximately 22% of methanogens are participate in the methylotrophic process. The archaea mainly involved in methylotrophy belong to the order Methanosarcinales, Methanococcoides, Methanosarcina, and Methanolobus and are the major methanogens involved in the methylotrophic process [57]. Compared to hydrogenotrophic methanogenesis, acetoclastic methanogenesis is less common in the rumen. The low abundance of these archaea may occur because their growth rate is slower than the acetate production rate [48]. The production of methane is primarily conducted by methanogenic archaea under anaerobic conditions. However, recent research has revealed that eukaryotes (including plants, animals, and fungi) can also actively participate in methane production in the presence of oxygen [58]. The methane emission strategies in the rumen target the above methanogens and the key enzymes in the methane production pathways. Balancing methane production and hydrogen retention is essential for achieving emission and normal fermentation by exploring the holistic function of the rumen microbial system.
Many microbial species involved in the above common functions within the rumen have been successfully cultured, which expands our knowledge on rumen biology and nutrition. These cultured species offer potential regulated targets to enhance the basic but important functions in the degradation of feed and contribute to the components of probiotics used for improving rumen digestion or feed digestibility. Based on the Hungate 1000 project, approximately 500 cultured rumen microbial species are currently available contributing to only 3.7% of the estimated total microbial numbers in the rumen [49,59]. Culture-independent approaches, such as meta-omics technologies are urgently needed to unravel the extensive functions in the rumen.
The concept of the metagenome was proposed by Handelsman et al [60] for the first time, and this concept refers to the sum of all microbial DNA in a specific environment. Metagenomics can sequence all microbial genetic material DNA in the sample, eliminating the problem that most microorganisms in the environment cannot be cultured [60]. Genes with potential functions in microorganisms can be obtained by open reading frame (ORF) prediction and functional annotation. Early research on rumen metagenomics mainly focused on exploring the genes encoding carbohydrate-active enzymes, especially the genes encoding lignocellulases that degrade plant cell walls. In 2009, Brulc et al [61] investigated carbohydrate-active enzymes in the rumen of three beef cattle fed the same diet. A total of 35 glycoside hydrolase genes were found, but only three carbohydrate-binding enzyme-encoding genes and three anchor modules were found. Although only three animals were used in this study, this study was the first to use metagenomic sequencing techniques to define a fiber adhesion microbial community. Later, metagenome sequencing was used to explore the functional genes of rumen microbial degradation by Hess et al [4]. The study increased the amount of metagenome sequencing data to 2.5 million ORFs, of which approximately 1% were identified as carbohydrate-active genes. Most of the genes were inconsistent with the NCBI nonredundant database, indicating that the rumen microbiome contains a wide range of fiber-degrading enzyme types. Ninety genes were selected for expression, 57% of which encoded enzymes with cellulolytic activity. In other ruminants, Pope et al [62] and Dai et al [63] reported the rumen metagenome of yaks and reindeer and also explored the genes encoding carbohydrate-active enzymes.
Metagenomic sequencing technology can also investigate the impact of methane inhibitors on rumen microbiota and their functionality, identifying key bacteria that regulate methane production. Ross et al [64] found that two distinct methane-reducing feed additives altered the microbial composition of samples in a similar manner, and from this result, they identified Faecalibacterium species as a potential biomarker for low methane-emitting cattle. Denman et al [65] discovered that inhibition of methanogens by bromochloromethane (BCM) directly and indirectly impacted the rumen microbiome. Among them, the relative abundance of hydrogen-utilizing bacteria such as Prevotella and Selenomonas species increased, resulting in the production of more propionate and suppression of methane generation. Later, a large number of studies on the methane emission of ruminants focused more on the natural selection of high- and low-methane emission animals using metagenomic technology. In beef cattle, methanogens and their genes are more abundant in the rumen of animals with high methane emissions [66,67]. Auffret et al [68] fed different diets to a group of beef cattle from different breeds to determine the rumen metagenome. It was found that the abundance of the methane generation pathway was strongly related to methane emissions, while the abundance of methanogens was weakly related to methane emissions. Shi et al [69] and Kamke et al [70] reported the relationship between rumen microbes and methane emission in goats using metagenomics. In addition to methane emissions, some metagenomics studies have also focused on beef cattle and cows [71,72], and reported the relationship between rumen microbial flora and function and feed utilization efficiency.
With the innovation and development of high-throughput sequencing technology and bioinformatics analysis tools, an increasing number of studies are using metagenome-assembled genomes (MAGs) for genomic analysis. In comparison to conventional metagenomic analysis, MAGs involve an additional step called metagenomic binning. This process involves categorizing the mixed sequences obtained from metagenomic sequencing or contigs assembled from the sequences into separate groups based on their respective species. MAGs with high completeness and low contamination levels were used to perform further taxonomic annotation and gene prediction [73]. MAGs are advantageous because they can overcome the limitations of reference genomes, including their availability and completeness. Metagenome assembly and binning can be performed de novo, enabling the discovery of new or uncultivable microorganisms [74]. Additionally, MAGs, through assembly, can identify short genes that might be missed by gene-finding tools; these short genes can be missed due to their small open reading frames (sORFs), which are a common feature of all genomes and hold significant untapped coding potential [75]. Due to these advantages, a limitation of conventional metagenomic analysis is addressed, making these tools broadly applicable to studies on the rumen microbiota.
As early as 2011, Hess et al [4] based on 268 G rumen metagenome data, successfully binned the genomes of 15 noncultivable microorganisms and verified them by single-cell whole-genome sequencing. Subsequently, researchers successively constructed the rumen MAGs of different ruminants. For example, 43 Scottish cattle, 913 bacterial and archaeal MAGs were assembled using more than 800 G of metagenomics data [5]; most of these strains had never been sequenced. In total, 69,000 proteins involved in carbohydrate metabolism were predicted, of which more than 90% had not been matched in the public database. Xie et al [76] first constructed a gene catalog of the entire gastrointestinal tract microbiota in ruminants, obtaining over 10,000 MAGs. This effort led to the identification of nearly 9,000 potentially novel bacteria and archaea, significantly expanding the known diversity and functions of the gastrointestinal microbiota in ruminant animals. MAGs are widely used to explore specific metabolic mechanisms of the rumen microbiota. Jiang et al [77] analyzed 17,000 gastrointestinal microbial genomes (10,373 MAGs from a previous study and 7,052 genomes from the collection of public ruminant microbial genomes). The researchers identified 2,366 high-quality genomes involved in the biosynthesis of vitamins B and K2. This study demonstrated regional heterogeneity and dietary effects on the potential for vitamin biosynthesis within the gastrointestinal microbiota of ruminant animals. In another study by Lin et al [78], they obtained 372 MAGs involved in bile acid (BA) transformation pathways were obtained from 108 samples of the entire gastrointestinal contents of 18 cows, revealing the rumen microbial BA metabolism mechanisms.
However, MAGs still have certain limitations, such as gaps, local assembly errors, chimeras, and contamination from other genomic fragments, which can restrict the value of these genomes [79]. These errors are often caused by immature sequencing technologies and bioinformatics algorithms. As sequencing depth increases and assembly techniques continue to improve, the quality of MAGs should be significantly improved. For instance, the high-quality sequencing technology of HiFi reads introduced by PacBio can enhance the completeness and accuracy of assembled genomes [80], and the development of assembly validation tools has played a crucial role in improving metagenome assembly [81]. Furthermore, there are still numerous uncharacterized microorganisms, and a comprehensive and well-curated reference gene database is needed for comparison and identification.
Although the rapid development of metagenomics technology has greatly enriched our knowledge on the rumen microbial diversity and function of ruminants, metagenomics still has some limitations. For example, metagenomics fails to reflect the real activity and functional characteristics of rumen microorganisms [82]. To investigate the composition of the active microbiota and the expression of active genes within the rumen microbiota at a specific time and space in situ, metatranscriptomics techniques are essential. Metatranscriptomics is a technique used to study the whole genome transcription and transcriptional regulation of microbial populations in a certain time and space, reflecting the true state of the rumen microbial community at the transcriptional level [83]. Compared with metagenomics, metatranscriptomics has been relatively slow to develop and has been applied to the study of rumen microorganisms with relatively few applications. A significant limitation is that RNA has a half-life period and tends to degrade during storage; thus, compared to total DNA, total RNA is much more challenging to extract total RNA than total DNA from rumen microorganisms.
Since most of the previous RNA sequencing analyses involved eukaryotic mRNA, the first large-scale rumen metatranscriptomics analysis also focused on rumen eukaryotes. Qi et al [84] used metatranscriptome technology to investigate the functional diversity of eukaryotic microorganisms within the rumen, revealing a significantly higher percentage of cellulase enzymes compared to metagenomics. The metatranscriptome can be a suitable approach to identify potential gene targets. Since then, an increasing number of studies on fiber degradation have employed metatranscriptomics technology. Dai et al [85] removed the rRNA from the total RNA and sequenced a total of 1 million nonrRNA sequences, of which approximately 1% were identified as carbohydrate-active enzymes or binding modules. In another cow study, similar levels of carbohydrate genes were obtained by Shinkai et al [86] using mRNA-enriched metatranscriptome sequencing data. The above studies confirmed that the main active bacteria responsible for fiber degradation were Fibrobacteraceae and Clostridiaceae. A cow study in 2017 used 18 new ribosome capture probes covering a large number of rumen archaea, bacteria, fungi, and prokaryotes, which confirmed that the bacteria could degrade fiber; in addition, fungi and protozoa greatly contributed to fiber degradation [87].
In terms of methane emission, the researchers used the rRNA and mcrA libraries to study rumen methanogens in the early stage and found a group of archaea similar to Thermoplasmatales. Because these microbes are ubiquitous in the rumen and encode mcrA genes, they may be methanogenic archaea [88]. However, before metatranscriptomics, a clear link between methanogenic capacity and these microorganisms could not be established. According to transcriptomics results, Poulsen identified a new cluster of methylotrophic methanogens [3]. In sheep, metatranscriptomics analysis revealed clear differences in rumen methanogens and related metabolic pathways between animals with low and high methane emissions, revealing a correlation between hydrogen trophic methanogens and methane production. However, based on metagenomics, no similar difference was found, which may result from the hydrogen supply of other rumen microbial fermentation pathways [69]. Following this study, metatranscriptomics was used to explore bacteria [70]. Based on this study, bacteria that were fermented into lactate and subsequently refermented into butyrate salts would decrease hydrogen production, thereby reducing methane generation.
Protein stands out as a direct and pivotal embodiment of microbial gene function. Therefore, research on protein composition and function based on the metagenome will help researchers study the abundance and distribution of functional molecules in microbial populations. Rumen metaproteomics describes the gene expression protein of the rumen microbial community in a specific time and space. Hart et al [89] compared the metaproteomics results with the protein functions predicted by metatranscriptomics and found that only 71% of the metaproteomics information matched the metatranscriptomics data, which indicated that metaproteomics could more accurately reflect the expression of environmentally active microorganisms than metatranscriptomics. Moreover, compared with the metagenome, the metaproteome is a more reliable indicator of animal phenotypes and achieves more accurate classifications [90]. As a result, metaproteomics plays a pivotal role in the study of microbial community function, and its advantages are attracting increasingly the attention of scientists. However, the method used to analyze metaproteomics bioinformatics still needs to be improved urgently. Due to the particularity of the research object, the analysis of metaproteomics requires a different set of bioinformatics and statistical models from those of traditional proteomics [91]. Therefore, biological meaning behind metaproteomics big data can only be determined once these problems are solved. With the increase and development of the next- and third-generation sequencing technology, and the significant reduction in sequencing costs, large-scale microbial sequence information is constantly being revealed, and the microbial genome database is also gradually improving; as a result, the protein identification method, speed, and accuracy of metaproteomics are promoted and will be significantly improved.
At present, there are relatively few applications of meta proteomics to rumen microorganisms. In an earlier study of rumen microbes in sheep, researchers attempted to identify cellulose-binding proteins through enrichment steps [92]. In this study, MS/MS-1D polyacrylamide gel electrophoresis (PAGE) was used to identify a small number of proteins and to link these proteins with microbial species. However, the limited database at that time restricted data mining. The proteins identified in this study include endoglucanase from F. succinogenes and exoglucanase from the fungus Piromyces equi. The combination of 2D PAGE separation technology and liquid chromatography-tandem mass spectrometry (LCŌĆōMS/MS) improves the resolution and facilitates the discovery of more peptides [93]. In this study, it was found that the enzyme of methanogens is among the most easily identified proteins, suggesting that metaproteomics may play an important role in exploring the rumen mechanism related to methane emission in ruminants. Recently, using metaproteomic analysis [94], the effects of digestion and methane metabolism in the rumen of ciliates have recently been elucidated. The shotgun metaproteome method can generate a much larger amount of data and offers a potential alternative to gel-based methods. Deusch and Seifert [95] were the first to employ the shotgun metaproteome method to identify prokaryotic and eukaryotic proteins in plant-attached microorganisms and rumen contents, showcasing a significant improvement in protein identification ratios. Subsequently, Deusch et al [96] conducted more complex explorations, identifying over 8,000 bacterial proteins and 350 archaeal proteins. These researchers also detected a substantial number of proteins involved in carbon metabolism.
Metagenomics, metatranscriptomics, and metaproteomics are used to study the life activities at the levels of genes, transcription, and proteins, respectively. Many of the biological activities in cells occur at the level of metabolites. For example, cell signal release, energy transmission, and intercellular communication are regulated by metabolites. Metabolomics is used to perform qualitative and quantitative analysis of all metabolites in an organism or a cell at a specific time and space [97]. The research objects of metabolomics are mostly small molecules with a relative molecular weight less than 1,000 [98]. Compared with metagenomics, metatranscriptomics, and metaproteomics, metabolomics can more easily detect and amplify small changes in gene and protein expression, making detection easier; in addition, stronger versatility and nonspecificity of metabolites are observed in various tissues. The results are more direct, and the metabolites can reflect the physiological and case status of the biological system. Targeted metabolomics determination commonly used in traditional ruminant nutrition research includes quantitative determination of a limited number of VFAs and quantitative determination of methane and hydrogen content [99,100]. Currently, nontargeted metabolic measurements are also widely used to study rumen microbial metabolomics [101], among which GCŌĆōMS and LCŌĆōMS are widely applied.
Metabolomics studies on ruminants often examine how the ratio of concentrate to roughage affects rumen metabolites, making it a significant area in ruminant metabolomics. Ametaj et al [101] and Saleem et al [100,102] conducted investigations on rumen metabolism changes with increasing proportions of concentrate in the diet. A variety of detection methods were used to identify rumen metabolites, and 246 of these metabolites were identified, mainly including phospholipids, inorganic ions, gases, amino acids, short-chain fatty acids, and carbohydrates. Combined with 87 metabolites reported in the literature, a database of rumen metabolites was created. In addition, other studies have reported changes in rumen metabolic patterns caused by changes in the ratio of concentrate to crude in the diet. For example, it was found that an increase in dietary cereals could lead to an increase in rumen methylamine content and a decrease in 3-phenylpropionate. Additionally, differences in the efficiency of feed utilization in ruminants can be reflected in rumen metabolites. The notable observation indicates the correlation between the metabolism of the rumen biological hydrogenation pathway (encompassing linoleic acid and alpha-linoleic acid) and average daily gain [99]. In cows, the increase in rumen short-chain fatty acid content and putrescine content and the decrease in methane content are related to high feed utilization efficiency [71].
Unlike other omics methods, metabolomics cannot directly link metabolites with microbial communities. Hence, the method must be combined with microbial relative abundance data obtained by other omics technologies for an integrated analysis. For example, in reports related to methane emissions, researchers found that changes in urine and plasma metabolites (trimethylamine N-oxide) were related to species of rumen protozoa and Methanomassilii cocus, indicating that trimethylamine N-oxide could be an important means of reflecting methane emissions [103,104]. In addition, the combined analysis of multiomics including rumen metabolomics effectively reflects the functional level information of nonculturable rumen bacteria. For example, by using metagenomics data to determine several genomes of the uncultured Bacteroides BS11 family, researchers subsequently validated their functions through metaproteomics and metabolomics data, which were identified it as crucial executors of hemicellulose degradation [105]. The combination of metabolomics with other omics technologies shows great potential to reveal mechanisms underlying ruminant diseases.
Research on the rumen microbiota has predominantly focused on rumen bacteria and archaea, but an increasing number of studies are focusing on other microorganisms present in the rumen. For example, rumen ciliates play a crucial role in the rumen environment, but their specific metabolic functions have remained unclear due to their nonculturable nature. Li et al [106] published the first catalog of rumen ciliate genomes, uncovering new ciliates and revealing their remarkable ability to degrade plant cell walls. Through the analysis of metaproteomic data from rumen samples, Andersen et al [94] provided a detailed description of specific metabolic niches occupied by ciliates in their microbiome environment, highlighting their significant impact on digestion and methane metabolism. In marine environments, viruses are driving factors in nutrient and energy cycling. However, research on the community of rumen viruses currently lags behind other topics. Using viral metagenome sequencing, Anderson et al [107] discovered that rumen viruses can breakdown complex carbohydrates and significantly influence microbial metabolism. Using proteomics, Solden et al [108] identified phages as active regulators of rumen ecosystem functionality. In terms of gene functionality, 50% to 70% of rumen viral reads possess viral replication capabilities, while other reads exhibit functional diversity. Auxiliary metabolic genes (AMGs) represent a subset of viral genes that redirect host metabolism toward reactions favorable for phage replication. Therefore, AMGs may reflect the potential impact of rumen viruses on microbial community metabolism. Nonetheless, there are still numerous unknown viral genes that must be further investigated. Omics technologies assist in gaining knowledge on viruses and their interactions. However, omics technologies still face some unresolved issues, such as limitations in the use of DNA sequencing methods to study RNA-based viruses and the predominance of phages in public reference databases, which potentially hinder the identification of archaeal viruses and others. Apart from ciliates and viruses, fungi and protists have emerged as new areas of research interest.
A multitude of emerging technologies are currently revolu tionizing our knowledge on microbiome functions. Organoids, which are self-organizing 3D tissues, can to mimic the intricate functions, structures, and biological complexity of organs. Specifically, intestinal organoids exhibit the complexity needed to replicate physiological and pathological conditions related to diet, microbiota, and host interactions, shedding light on the mechanisms governing microbial-related functions [109]. The concept of the ŌĆ£RamanomeŌĆØ involves compiling multiple single-cell Raman spectra from cell populations at specific conditions and time points. As demonstrated by Jing et al [110], the Ramanome approach has been successfully utilized to identify specific functional bacteria isolated from seawater. Raman spectroscopy, through which individual microbial cells can be analyzed in a nondestructive manner, allows for further cell cultivation or DNA analysis, ultimately improving our knowledge on their functional activities. Through applying these emerging technologies, we can gain comprehensive insights into the intricate roles of microorganisms, making it possible to effectively target and regulate the physiological functions of animal organisms by manipulating the rumen microbiota.
Through metagenomics and other techniques, we have clarified the functionality of rumen microbiota to further manipulate them. Santra et al [111] suggested that digestive functions can be enhanced and nutrition and productivity can be optimized by manipulating these microorganisms. Currently, various methods are used to manipulate rumen microbiota, which occurs primarily through dietary control, such as the use of chemical additives, direct microbial supplementation, and probiotics [112]. However, the effects of dietary control on the adult rumen microbiome and fermentation are typically effects. Additionally, some methods involve the transfection of rumen microbiota, in which microbial communities are physically introduced into the cowŌĆÖs rumen; however, these transplanted microorganisms do not seem to persist [54]. Therefore, extensive research is still needed to clarify the functionality of the rumen microbiota and determine how to manipulate it effectively. Current approaches primarily focus on manipulating heritable microorganisms through breeding programs, as these microorganisms are vital constituents of the symbiotic network within the rumen microbiota, potentially exerting pleiotropic effects on microbial composition [113]. Another approach is early-life interventions, which may generate long-term effects on rumen function [114]. However, the optimal time for producing sustained effects by manipulating the rumen microbiota remains to be determined.
Through the collective action of the rumen microbiota, high-fiber feed that monogastric animals cannot digest can be degraded and fermented. This process serves as an energy source and a microbial protein source for ruminant animals. However, it also results in the production of significant amounts of methane. Numerous omics technologies have been employed to characterize the rumen microbiota, continuously seeking fresh insights into the functionality of these complex microbial communities. Partitioning microbial communities into functional groups based on metabolic functions provides a more precise assessment of the communityŌĆÖs status and functions, addressing limitations associated with exclusive reliance on species taxonomy, including vagueness and redundancy. Integrating taxonomy with functional groups enhances our comprehensive understanding of the rumen ecosystemŌĆÖs functions, thereby facilitating the design of interventions within the rumen microbiota. As a result, the performance of the rumen ecosystem can be further improved.
Hence, through exploring microbial communities more comprehensively with advanced and innovative omics techniques, the rumen microbiota can be more easily manipulated through diet or other means, improving the production efficiency of ruminant animals, reducing methane emissions, and providing more possibilities for the future of sustainable livestock farming.
Notes
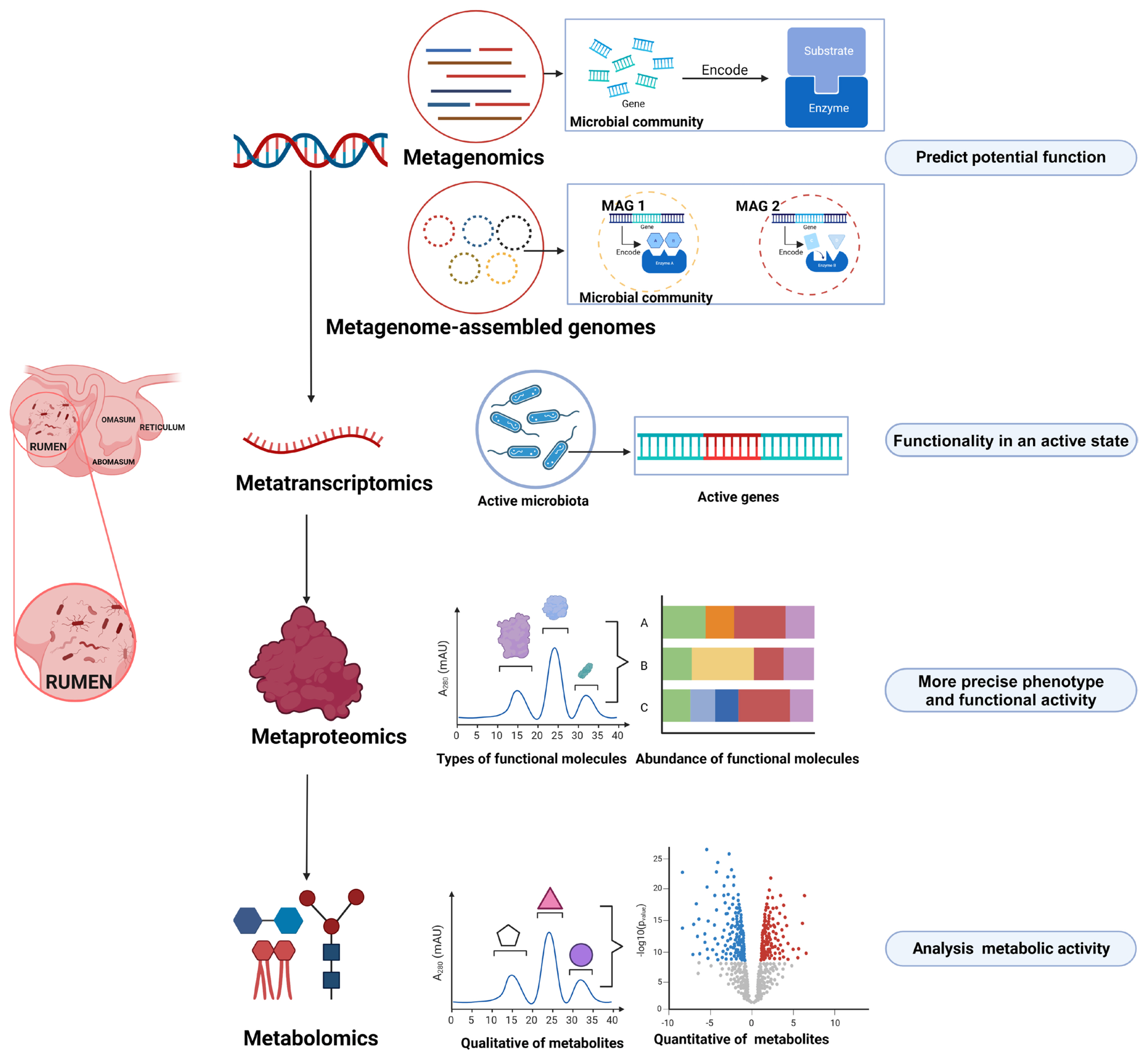
Figure┬Ā1
Omics technologies for investigating rumen microbial function (Created with BioRender.com).
Table┬Ā1
Classification of major rumen functional bacteria
| Functional classification | Species | Reference |
|---|---|---|
| Cellulolytic bacteria | Fibrobacter succinogenes, Ruminococcus flavefaciens, Ruminococcus albus, Clostridium lochheadii, Clostridium longisporum, Butyrivibrio fibrisolvens, Eubacterium cellulosolvens | [12], [14], [15] |
| Hemicellulolytic bacteria | Cellulolytic bacteria, Lachnospira multiparus, Prevotella sp., Pseudobutyrivibrio xylanivorans, Streptococcus sp. | [19], [20] |
| Amylolytic bacteria | Fibrobacter succinogenes, Streptococcus bovis, Prevotella ruminicola, Clostridium sp., Ruminobacter amylophilus, Succinimonas amylolytica, Ruminococcus bromii, Selenomonas ruminantium, Butyrivibrio fibrisolvens | [22], [23], [24] |
| Pectin-degrading bacteria | Fibrobacter succinogenes, Ruminococcus albus, Butyrivibrio fibrisolvens, Streptococcus bovis, Treponema sp., Cellulosilyticum ruminicola, Lachnospira multipara, Prevotella sp. | [30], [31], [32] |
| Proteolytic bacteria | Butyrivibrio fibrisolvens, Butyrivibrio proteoclasticus, Streptococcus bovis, Prevotella sp., Ruminobacter amylophilus, Eubacterium budayi, Selenomonas ruminantium | [34], [36], [37] |
| Peptidolytic bacteria | Fibrobacter succinogenes, Prevotella sp., Ruminococcus sp., Eubacterium ruminantium, Streptococcus bovis, Ruminobacter amylophilus, Veillonella parvula, Megasphaera elsdenii, Lachnospira multipara | [37], [41] |
| Lipid-degrading bacteria | Anaerovibrio lipolytica, Butyrivibrio fibrisolvens, Clostridium sp., Propionibacterium sp. | [43], [44] |
| Biohydrogenating bacteria | Butyrivibrio hungatei, Butyrivibrio proteoclasticus, Propionibacterium acnes, Eubacterium ruminantium, Clostridium proteoclasticum, Pseudobutyrivibrio sp. | [44], [48] |
| Lactic producing bacteria | Bifidobacterium lactis, Lactobacillus acidophilus, Streptococcus bovis | [52] |
| Lactic utilising bacteria | Selenomonas ruminantium, Megasphaera elsdenii | [52] |
| Succinate producing bacteria | Actinobacillus succinogenes, Mannheimia succiniciproducens | [50] |
| Succinat utilising bacteria | Succiniclasticum ruminis | [51] |
| Deaminating bacteria | Butyrivibrio fibrisolvens, Prevotella ruminicola, Megasphaera elsdenii, Allisonella histaminiformans, Clostridium aminophilum, Clostridium sticklandii, Peptostreptococcus anaerobius | [53] |
| Glycerol fermenting bacteria | Anaerovibrio lipolytica, Butyrivibrio fibrisolvens, Selenomonas ruminantium | [48] |
REFERENCES
1. Langda S, Zhang C, Zhang K, et al. Diversity and composition of rumen bacteria, fungi, and protozoa in goats and sheep living in the same high-altitude pasture. Animals (Basel) 2020; 10:186
https://doi.org/10.3390/ani10020186



2. Cammack KM, Austin KJ, Lamberson WR, Conant GC, Cunningham HC. RUMINNAT NUTRITION SYMPOSIUM: Tiny but mighty: the role of the rumen microbes in livestock production. J Anim Sci 2018; 96:752ŌĆō70.
https://doi.org/10.1093/jas/skx053



3. Poulsen M, Schwab C, Borg Jensen B, et al. Methylotrophic methanogenic Thermoplasmata implicated in reduced methane emissions from bovine rumen. Nat Commun 2013; 4:1428
https://doi.org/10.1038/ncomms2432


4. Hess M, Sczyrba A, Egan R, et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 2011; 331:463ŌĆō7.
https://doi.org/10.1126/science.1200387


5. Stewart RD, Auffret MD, Warr A, et al. Assembly of 913 microbial genomes from metagenomic sequencing of the cow rumen. Nat Commun 2018; 9:870
https://doi.org/10.1038/s41467-018-03317-6



6. Mora├»s S, Mizrahi I. The road not taken: the rumen microbiome, functional groups, and community states. Trends Microbiol 2019; 27:538ŌĆō49.
https://doi.org/10.1016/j.tim.2018.12.011


7. Benson AK, Kelly SA, Legge R, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci USA 2010; 107:18933ŌĆō8.
https://doi.org/10.1073/pnas.1007028107



8. Taxis TM, Wolff S, Gregg SJ, et al. The players may change but the game remains: network analyses of ruminal microbiomes suggest taxonomic differences mask functional similarity. Nucleic Acids Res 2015; 43:9600ŌĆō12.
https://doi.org/10.1093/nar/gkv973



9. Boon E, Meehan CJ, Whidden C, Wong DHJ, Langille MGI, Beiko RG. Interactions in the microbiome: communities of organisms and communities of genes. FEMS Microbiol Rev 2020; 38:90ŌĆō118.
https://doi.org/10.1111/1574-6976.12035



10. Ransom-Jones E, Jones DL, McCarthy AJ, McDonald JE. The fibrobacteres: an important phylum of cellulose-degrading bacteria. Microb Ecol 2012; 63:267ŌĆō81.
https://doi.org/10.1007/s00248-011-9998-1


11. Mora├»s S, Mizrahi I. Islands in the stream: from individual to communal fiber degradation in the rumen ecosystem. FEMS Microbiol Rev 2019; 43:362ŌĆō79.
https://doi.org/10.1093/femsre/fuz007



12. Stevenson DM, Weimer PJ. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl Microbiol Biotechnol 2007; 75:165ŌĆō74.
https://doi.org/10.1007/s00253-006-0802-y


13. Yeoman CJ, Fields CJ, Lepercq P, et al. In vivo competitions between fibrobacter succinogenes, ruminococcus flavefaciens, and ruminoccus albus in a gnotobiotic sheep model revealed by multi-omic analyses. Mbio 2021; 12:10.1128/mbio.03533.20
https://doi.org/10.1128/mbio.03533-20

14. Hungate RE. Microorganisms in the rumen of cattle fed a constant ration. Can J Microbiol 1957; 3:289ŌĆō311.
https://doi.org/10.1139/m57-034


15. Rodr├Łguez Hern├Īez J, Cer├│n Cucchi ME, Cravero S, et al. The first complete genomic structure of Butyrivibrio fibrisolvens and its chromid. Microb Genomics 2018; 4:e000216
https://doi.org/10.1099/mgen.0.000216

16. Weimer PJ. Degradation of cellulose and hemicellulose by ruminal microorganisms. Microorganisms 2022; 10:2345
https://doi.org/10.3390/microorganisms10122345



17. Paul SS, Deb SM, Punia BS, Singh D, Kumar R. Fibrolytic potential of anaerobic fungi (Piromyces sp.) isolated from wild cattle and blue bulls in pure culture and effect of their addition on in vitro fermentation of wheat straw and methane emission by rumen fluid of buffaloes. J Sci Food Agric 2010; 90:1218ŌĆō26.
https://doi.org/10.1002/jsfa.3952


18. Saye LMG, Navaratna TA, Chong JPJ, OŌĆÖMalley MA, Theodorou MK, Reilly M. The anaerobic fungi: challenges and opportunities for industrial lignocellulosic biofuel production. Microorganisms 2021; 9:694
https://doi.org/10.3390/microorganisms9040694



19. Coen JA, Dehority BA. Degradation and utilization of hemicellulose from intact forages by pure cultures of rumen bacteria. Appl Microbiol 1970; 20:362ŌĆō8.
https://doi.org/10.1128/am.20.3.362-368.1970



20. Emerson EL, Weimer PJ. Fermentation of model hemicelluloses by Prevotella strains and Butyrivibrio fibrisolvens in pure culture and in ruminal enrichment cultures. Appl Microbiol Biotechnol 2017; 101:4269ŌĆō78.
https://doi.org/10.1007/s00253-017-8150-7


21. Patel S, Ambalam P. Role of rumen protozoa: metabolic and fibrolytic. Adv Biotechnol Microbiol 2018; 10:555793
https://doi.org/10.19080/AIBM.2018.10.555793

22. Anderson KL. Biochemical analysis of starch degradation by Ruminobacter amylophilus 70. Appl Environ Microbiol 1995; 61:1488ŌĆō91.
https://doi.org/10.1128/aem.61.4.1488-1491.1995



23. Cerqueira FM, Photenhauer AL, Pollet RM, Brown HA, Koropatkin NM. Starch digestion by gut bacteria: crowdsourcing for carbs. Trends Microbiol 2020; 28:95ŌĆō108.
https://doi.org/10.1016/j.tim.2019.09.004


24. Ze X, Ben David Y, Laverde-Gomez JA, et al. Unique organization of extracellular amylases into amylosomes in the resistant starch-utilizing human colonic firmicutes bacterium Ruminococcus bromii. Mbio 2015; 6:e01058ŌĆō01015.
https://doi.org/10.1128/mBio.01058-15



25. Hua D, Hendriks WH, Xiong B, Pellikaan WF. Starch and cellulose degradation in the rumen and applications of metagenomics on ruminal microorganisms. Animals-Basel 2022; 12:3020
https://doi.org/10.3390/ani12213020



26. McAllister TA, Cheng KJ, Rode LM, Forsberg CW. Digestion of barley, maize, and wheat by selected species of ruminal bacteria. Appl Environ Microbiol 1990; 56:3146ŌĆō53.
https://doi.org/10.1128/aem.56.10.3146-3153.1990



27. McAllister TA, Cheng KJ. Microbial strategies in the ruminal digestion of cereal grains. Anim Feed Sci Technol 1996; 62:29ŌĆō36.
https://doi.org/10.1016/S0377-8401(96)01003-6

28. Coleman GS. The metabolism of rumen ciliate protozoa. FEMS Microbiol Rev 1986; 2:321ŌĆō44.
https://doi.org/10.1111/j.1574-6968.1986.tb01864.x

29. McAllister TA, Dong Y, Yank LJ, et al. Cereal grain digestion by selected strains of ruminal fungi. Can J Microbiol 1993; 39:367ŌĆō76.
https://doi.org/10.1139/m93-054


30. Liu J, Wang JK, Zhu W, et al. Monitoring the rumen pectinolytic bacteria Treponema saccharophilum using real-time PCR. FEMS Microbiol Ecol 2014; 87:576ŌĆō85.
https://doi.org/10.1111/1574-6941.12246


31. Cai S, Li J, Hu FZ, et al. Cellulosilyticum ruminicola, a newly described rumen bacterium that possesses redundant fibrolytic-protein-encoding genes and degrades lignocellulose with multiple carbohydrate-borne fibrolytic enzymes. Appl Environ Microbiol 2010; 76:3818ŌĆō24.
https://doi.org/10.1128/AEM.03124-09



32. Marounek M, Du┼Īkov├Ī D. Metabolism of pectin in rumen bacteria Butyrivibrio fibrisolvens and Prevotella ruminicola. Lett Appl Microbiol 1999; 29:429ŌĆō33.
https://doi.org/10.1046/j.1472-765X.1999.00671.x

33. Tan P, Liu H, Zhao J, et al. Amino acids metabolism by rumen microorganisms: Nutrition and ecology strategies to reduce nitrogen emissions from the inside to the outside. Sci Total Environ 2021; 800:149596
https://doi.org/10.1016/j.scitotenv.2021.149596


34. Hartinger T, Gresner N, S├╝dekum KH. Does intra-ruminal nitrogen recycling waste valuable resources? A review of major players and their manipulation. J Anim Sci Biotechnol 2018; 9:33
https://doi.org/10.1186/s40104-018-0249-x



35. Belanche A, Doreau M, Edwards JE, Moorby JM, Pinloche E, Newbold CJ. Shifts in the rumen microbiota due to the type of carbohydrate and level of protein ingested by dairy cattle are associated with changes in rumen fermentation. J Nutr 2012; 142:1684ŌĆō92.
https://doi.org/10.3945/jn.112.159574


36. Cotta MA, Hespell RB. Proteolytic activity of the ruminal bacterium Butyrivibrio fibrisolvens. Appl Environ Microbiol 1986; 52:51ŌĆō8.
https://doi.org/10.1128/aem.52.1.51-58.1986



37. Wallace RJ, McKain N, Broderick GA, et al. Peptidases of the rumen bacterium, Prevotella ruminicola. Anaerobe 1997; 3:35ŌĆō42.
https://doi.org/10.1006/anae.1996.0065


38. Prins RA, van Rheenen DL, vanŌĆÖt Klooster AT. Characterization of microbial proteolytic enzymes in the rumen. Antonie van Leeuwenhoek 1983; 49:585ŌĆō95.
https://doi.org/10.1007/BF00399852


39. Coleman GS. Hydrolysis of Fraction 1 leaf protein and casein by rumen entodiniomorphid protozoa. J Appl Bacteriol 1983; 55:111ŌĆō8.
https://doi.org/10.1111/j.1365-2672.1983.tb02654.x

40. Asao N, Ushida K, Kojima Y. Proteolytic activity of rumen fungi belonging to the genera Neocallimastix and Piromyces. Lett Appl Microbiol 1993; 16:247ŌĆō50.
https://doi.org/10.1111/j.1472-765X.1993.tb01410.x

41. Wallace RJ, McKain N. A survey of peptidase activity in rumen bacteria. J Gen Microbiol 1991; 137:2259ŌĆō64.
https://doi.org/10.1099/00221287-137-9-2259


42. Wallace RJ, McKain N, Newbold CJ. Metabolism of small peptides in rumen fluid. Accumulation of intermediates during hydrolysis of alanine oligomers, and comparison of peptidolytic activities of bacteria and protozoa. J Sci Food Agric 1990; 50:191ŌĆō9.
https://doi.org/10.1002/jsfa.2740500207

43. Hobson PN, Mann SO. The isolation of glycerol-fermenting and lipolytic bacteria from the rumen of the sheep. J Gen Microbiol 1961; 25:227ŌĆō40.
https://doi.org/10.1099/00221287-25-2-227


44. Jarvis GN, Moore ERB. Lipid metabolism and the rumen microbial ecosystem. Timmis KN, editorHandbook of hydrocarbon and lipid microbiology. Berlin, Heidelberg, Germany: Springer; 2010. p. 2245ŌĆō57.
https://doi.org/10.1007/978-3-540-77587-4_163

45. Wallace RJ, Brammall ML. The role of different species of bacteria in the hydrolysis of protein in the rumen. J Gen Microbiol 1985; 131:821ŌĆō32.
https://doi.org/10.1099/00221287-131-4-821

46. Louren├¦o M, Ramos-Morales E, Wallace RJ. The role of microbes in rumen lipolysis and biohydrogenation and their manipulation. Animal 2010; 4:1008ŌĆō23.
https://doi.org/10.1017/S175173111000042X


47. Jenkins TC, Wallace RJ, Moate PJ, Mosley EE. BOARD-INVITED REVIEW: Recent advances in biohydrogenation of unsaturated fatty acids within the rumen microbial ecosystem. J Anim Sci 2008; 86:397ŌĆō412.
https://doi.org/10.2527/jas.2007-0588


48. Nagaraja TG. Microbiology of the rumen. Millen DD, De Beni Arrigoni M, Lauritano Pacheco RD, editorsRumenology. Cham: Springer International Publishing; 2016. p. 39ŌĆō61.
https://doi.org/10.1007/978-3-319-30533-2_2

49. Seshadri R, Leahy SC, Attwood GT, et al. Cultivation and sequencing of rumen microbiome members from the Hungate 1000 collection. Nat Biotechnol 2018; 36:359ŌĆō67.
https://doi.org/10.1038/nbt.4110



50. Song H, Lee SY. Production of succinic acid by bacterial fermentation. Enzyme Microb Technol 2006; 39:352ŌĆō61.
https://doi.org/10.1016/j.enzmictec.2005.11.043

51. Gylswyk van NO. Succiniclasticum ruminis gen. nov., sp. nov., a ruminal bacterium converting succinate to propionate as the sole energy-yielding mechanism. Int J Syst Evol Microbiol 1995; 45:297ŌĆō300.
https://doi.org/10.1099/00207713-45-2-297

52. Wei W, Zhen Y, Wang Y, Shahzad K, Wang M. Advances of rumen functional bacteria and the application of micro-encapsulation fermentation technology in ruminants: a review. Fermentation 2022; 8:564
https://doi.org/10.3390/fermentation8100564

53. Wallace RJ. Ruminal microbial metabolism of peptides and amino acids. J Nutr 1996; 126:1326SŌĆō34S.
https://doi.org/10.1093/jn/126.suppl_4.1326S


54. Mizrahi I, Wallace RJ, Mora├»s S. The rumen microbiome: balancing food security and environmental impacts. Nat Rev Microbiol 2021; 19:553ŌĆō66.
https://doi.org/10.1038/s41579-021-00543-6


55. Malik PK, Trivedi S, Mohapatra A, et al. Comparison of enteric methane yield and diversity of ruminal methanogens in cattle and buffaloes fed on the same diet. PLoS One 2021; 16:e0256048
https://doi.org/10.1371/journal.pone.0256048



56. Lan W, Yang C. Ruminal methane production: associated microorganisms and the potential of applying hydrogen-utilizing bacteria for mitigation. Sci Total Environ 2019; 654:1270ŌĆō83.
https://doi.org/10.1016/j.scitotenv.2018.11.180


57. Liu Y, Whitman WB. Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann NY Acad Sci 2008; 1125:171ŌĆō89.
https://doi.org/10.1196/annals.1419.019


58. Liu J, Chen H, Zhu Q, et al. A novel pathway of direct methane production and emission by eukaryotes including plants, animals and fungi: an overview. Atmos Environ 2015; 115:26ŌĆō35.
https://doi.org/10.1016/j.atmosenv.2015.05.019

59. Watson M. New insights from 33,813 publicly available metagenome-assembled-genomes (MAGs) assembled from the rumen microbiome. bioRxiv 2021; 2021.04.02.438222https://doi.org/10.1101/2021.04.02.438222

60. Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol 1998; 5:R245ŌĆō9.
https://doi.org/10.1016/s1074-5521(98)90108-9


61. Brulc JM, Antonopoulos DA, Berg Miller ME, et al. Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc Natl Acad Sci USA 2009; 106:1948ŌĆō53.
https://doi.org/10.1073/pnas.0806191105



62. Pope PB, Mackenzie AK, Gregor I, et al. Metagenomics of the svalbard reindeer rumen microbiome reveals abundance of polysaccharide utilization loci. PLoS One 2012; 7:e38571
https://doi.org/10.1371/journal.pone.0038571



63. Dai X, Zhu Y, Luo Y, et al. Metagenomic insights into the fibrolytic microbiome in yak rumen. PLoS One 2012; 7:e40430
https://doi.org/10.1371/journal.pone.0040430



64. Ross EM, Moate PJ, Marett L, Cocks BG, Hayes BJ. Investigating the effect of two methane-mitigating diets on the rumen microbiome using massively parallel sequencing. J Dairy Sci 2013; 96:6030ŌĆō46.
https://doi.org/10.3168/jds.2013-6766


65. Denman SE, Martinez Fernandez G, Shinkai T, Mitsumori M, McSweeney CS. Metagenomic analysis of the rumen microbial community following inhibition of methane formation by a halogenated methane analog. Front Microbiol 2015; 6:1087
https://doi.org/10.3389/fmicb.2015.01087



66. Auffret MD, Dewhurst RJ, Duthie CA, et al. The rumen microbiome as a reservoir of antimicrobial resistance and pathogenicity genes is directly affected by diet in beef cattle. Microbiome 2017; 5:159
https://doi.org/10.1186/s40168-017-0378-z



67. Wallace RJ, Rooke JA, McKain N, et al. The rumen microbial metagenome associated with high methane production in cattle. BMC Genomics 2015; 16:839
https://doi.org/10.1186/s12864-015-2032-0



68. Auffret MD, Stewart R, Dewhurst RJ, et al. Identification, comparison, and validation of robust rumen microbial biomarkers for methane emissions using diverse bos taurus breeds and basal diets. Front Microbiol 2018; 8:2642
https://doi.org/10.3389/fmicb.2017.02642



69. Shi W, Moon CD, Leahy SC, et al. Methane yield phenotypes linked to differential gene expression in the sheep rumen microbiome. Genome Res 2014; 24:1517ŌĆō25.
https://doi.org/10.1101/gr.168245.113



70. Kamke J, Kittelmann S, Soni P, et al. Rumen metagenome and metatranscriptome analyses of low methane yield sheep reveals a Sharpea-enriched microbiome characterised by lactic acid formation and utilisation. Microbiome 2016; 4:56
https://doi.org/10.1186/s40168-016-0201-2



71. Shabat SKB, Sasson G, Doronfaigenboim A, et al. Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J 2016; 10:2958ŌĆō72.
https://doi.org/10.1038/ismej.2016.62



72. Li F, Hitch TCA, Chen Y, Creevey CJ, Guan LL. Comparative metagenomic and metatranscriptomic analyses reveal the breed effect on the rumen microbiome and its associations with feed efficiency in beef cattle. Microbiome 2019; 7:6
https://doi.org/10.1186/s40168-019-0618-5



73. Yang C, Chowdhury D, Zhang Z, et al. A review of computational tools for generating metagenome-assembled genomes from metagenomic sequencing data. Comput Struct Biotechnol J 2021; 19:6301ŌĆō14.
https://doi.org/10.1016/j.csbj.2021.11.028



74. Meijenfeldt von FAB, Arkhipova K, Cambuy DD, Coutinho FH, Dutilh BE. Robust taxonomic classification of uncharted microbial sequences and bins with CAT and BAT. Genome Biol 2019; 20:217
https://doi.org/10.1186/s13059-019-1817-x



75. Andrews SJ, Rothnagel JA. Emerging evidence for functional peptides encoded by short open reading frames. Nat Rev Genet 2014; 15:193ŌĆō204.
https://doi.org/10.1038/nrg3520


76. Xie F, Jin W, Si H, et al. An integrated gene catalog and over 10,000 metagenome-assembled genomes from the gastrointestinal microbiome of ruminants. Microbiome 2021; 9:137
https://doi.org/10.1186/s40168-021-01078-x



77. Jiang Q, Lin L, Xie F, et al. Metagenomic insights into the microbe-mediated B and K2 vitamin biosynthesis in the gastrointestinal microbiome of ruminants. Microbiome 2022; 10:109
https://doi.org/10.1186/s40168-022-01298-9



78. Lin L, Lai Z, Yang H, et al. Genome-centric investigation of bile acid metabolizing microbiota of dairy cows and associated diet-induced functional implications. ISME J 2023; 17:172ŌĆō84.
https://doi.org/10.1038/s41396-022-01333-5



79. Chen LX, Anantharaman K, Shaiber A, Murat Eren A, Banfield JF. Accurate and complete genomes from metagenomes. Genome Res 2020; 30:315ŌĆō33.
https://doi.org/10.1101/gr.258640.119



80. Kim CY, Ma J, Lee I. HiFi metagenomic sequencing enables assembly of accurate and complete genomes from human gut microbiota. Nat Commun 2022; 13:6367
https://doi.org/10.1038/s41467-022-34149-0



81. Olson ND, Treangen TJ, Hill CM, et al. Metagenomic assembly through the lens of validation: recent advances in assessing and improving the quality of genomes assembled from metagenomes. Brief Bioinform 2019; 20:1140ŌĆō50.
https://doi.org/10.1093/bib/bbx098



82. Simon C, Daniel R. Metagenomic analyses: past and future trends. Appl Environ Microbiol 2011; 77:1153ŌĆō61.
https://doi.org/10.1128/AEM.02345-10



83. Aguiar-Pulido V, Huang W, Suarez-Ulloa V, Cickovski T, Mathee K, Narasimhan G. Metagenomics, metatranscriptomics, and metabolomics approaches for microbiome analysis. Evol Bioinform Online 2016; 12s1:5ŌĆō16.
https://doi.org/10.4137/EBO.S36436



84. Qi M, Wang P, OŌĆÖToole N, et al. Snapshot of the eukaryotic gene expression in muskoxen rumenŌĆöa metatranscriptomic approach. PLoS One 2011; 6:e20521
https://doi.org/10.1371/journal.pone.0020521



85. Dai X, Tian Y, Li J, et al. Metatranscriptomic analyses of plant cell wall polysaccharide degradation by microorganisms in the cow rumen. Appl Environ Microbiol 2015; 81:1375ŌĆō86.
https://doi.org/10.1128/AEM.03682-14



86. Shinkai T, Mitsumori M, Sofyan A, et al. Comprehensive detection of bacterial carbohydrate-active enzyme coding genes expressed in cow rumen. Anim Sci J 2016; 87:1363ŌĆō70.
https://doi.org/10.1111/asj.12585


87. Comtet-Marre S, Parisot N, Lepercq P, et al. Metatranscriptomics reveals the active bacterial and eukaryotic fibrolytic communities in the rumen of dairy cow fed a mixed diet. Front Microbiol 2017; 8:67
https://doi.org/10.3389/fmicb.2017.00067



88. Denman SE, Tomkins NW, McSweeney CS. Quantitation and diversity analysis of ruminal methanogenic populations in response to the antimethanogenic compound bromochloromethane. FEMS Microbiol Ecol 2007; 62:313ŌĆō22.
https://doi.org/10.1111/j.1574-6941.2007.00394.x


89. Hart EH, Creevey CJ, Hitch T, Kingston-Smith AH. Meta-proteomics of rumen microbiota indicates niche compartmentalisation and functional dominance in a limited number of metabolic pathways between abundant bacteria. Sci Rep 2018; 8:10504
https://doi.org/10.1038/s41598-018-28827-7



90. Sasson G, Mora├»s S, Kokou F, et al. Metaproteome plasticity sheds light on the ecology of the rumen microbiome and its connection to host traits. ISME J 2022; 16:2610ŌĆō21.
https://doi.org/10.1038/s41396-022-01295-8



91. Schiebenhoefer H, Van Den Bossche T, Fuchs S, Renard BY, Muth T, Martens L. Challenges and promise at the interface of metaproteomics and genomics: an overview of recent progress in metaproteogenomic data analysis. Expert Rev Proteomics 2019; 16:375ŌĆō90.
https://doi.org/10.1080/14789450.2019.1609944


92. Toyoda A, Iio W, Mitsumori M, et al. Isolation and identification of cellulose-binding proteins from sheep rumen contents. Appl Environ Microbiol 2009; 75:1667ŌĆō73.
https://doi.org/10.1128/aem.01838-08



93. Snelling TJ, Wallace RJ. The rumen microbial metaproteome as revealed by SDS-PAGE. BMC Microbiol 2017; 17:9
https://doi.org/10.1186/s12866-016-0917-y



94. Andersen TO, Altshuler I, Vera-Ponce de Le├│n A, et al. Metabolic influence of core ciliates within the rumen microbiome. ISME J 2023; 17:1128ŌĆō40.
https://doi.org/10.1038/s41396-023-01407-y



95. Deusch S, Seifert J. Catching the tip of the iceberg ŌĆō Evaluation of sample preparation protocols for metaproteomic studies of the rumen microbiota. Proteomics 2015; 15:3590ŌĆō5.
https://doi.org/10.1002/pmic.201400556


96. Deusch S, Camarinha-Silva A, Conrad J, Beifuss U. A structural and functional elucidation of the rumen microbiome influenced by various diets and microenvironments. Front Microbiol 2017; 8:1605
https://doi.org/10.3389/fmicb.2017.01605



97. Kueger S, Steinhauser D, Willmitzer L, Giavalisco P. High-resolution plant metabolomics: from mass spectral features to metabolites and from whole-cell analysis to subcellular metabolite distributions. Plant J 2012; 70:39ŌĆō50.
https://doi.org/10.1111/j.1365-313X.2012.04902.x


98. Yang Q, Zhang A, Miao J, et al. Metabolomics biotechnology, applications, and future trends: a systematic review. RSC Adv 2019; 9:37245ŌĆō57.
https://doi.org/10.1039/C9RA06697G



99. Artegoitia VM, Foote AP, Lewis RM, Freetly HC. Rumen fluid metabolomics analysis associated with feed efficiency on crossbred steers. Sci Rep 2017; 7:2864
https://doi.org/10.1038/s41598-017-02856-0



100. Saleem F, Ametaj BN, Bouatra S, et al. A metabolomics approach to uncover the effects of grain diets on rumen health in dairy cows. J Dairy Sci 2012; 95:6606ŌĆō23.
https://doi.org/10.3168/jds.2012-5403


101. Ametaj BN, Zebeli Q, Saleem F, et al. Metabolomics reveals unhealthy alterations in rumen metabolism with increased proportion of cereal grain in the diet of dairy cows. Metabolomics 2010; 6:583ŌĆō94.
https://doi.org/10.1007/s11306-010-0227-6

102. Saleem F, Bouatra S, Guo AC, et al. The bovine ruminal fluid metabolome. Metabolomics 2013; 9:360ŌĆō78.
https://doi.org/10.1007/s11306-012-0458-9

103. Saro C, Hohenester UM, Bernard M, et al. Effectiveness of interventions to modulate the rumen microbiota composition and function in pre-ruminant and ruminant lambs. Front Microbiol 2018; 9:1273
https://doi.org/10.3389/fmicb.2018.01273



104. Morgavi DP, Rathahao-Paris E, Popova M, Boccard J, Nielsen KF, Boudra H. Rumen microbial communities influence metabolic phenotypes in lambs. Front Microbiol 2015; 6:1060
https://doi.org/10.3389/fmicb.2015.01060



105. Solden LM, Hoyt DW, Collins WB, et al. New roles in hemicellulosic sugar fermentation for the uncultivated Bacteroidetes family BS11. ISME J 2017; 11:691ŌĆō703.
https://doi.org/10.1038/ismej.2016.150



106. Li Z, Wang X, Zhang Y, et al. Genomic insights into the phylogeny and biomass-degrading enzymes of rumen ciliates. ISME J 2022; 16:2775ŌĆō87.
https://doi.org/10.1038/s41396-022-01306-8



107. Anderson CL, Sullivan MB, Fernando SC. Dietary energy drives the dynamic response of bovine rumen viral communities. Microbiome 2017; 5:155
https://doi.org/10.1186/s40168-017-0374-3



108. Solden LM, Naas AE, Roux S, et al. Interspecies cross-feeding orchestrates carbon degradation in the rumen ecosystem. Nat Microbiol 2018; 3:1274ŌĆō84.
https://doi.org/10.1038/s41564-018-0225-4



109. Kadosh E, Snir-Alkalay I, Venkatachalam A, et al. The gut microbiome switches mutant p53 from tumour-suppressive to oncogenic. Nature 2020; 586:133ŌĆō8.
https://doi.org/10.1038/s41586-020-2541-0



110. Jing X, Gou H, Gong Y, et al. Raman-activated cell sorting and metagenomic sequencing revealing carbon-fixing bacteria in the ocean. Environ Microbiol 2018; 20:2241ŌĆō55.
https://doi.org/10.1111/1462-2920.14268



111. Santra A, Karim SA. Rumen manipulation to improve animal productivity. Asian-Australas J Anim Sci 2003; 16:748ŌĆō63.
https://doi.org/10.5713/ajas.2003.748

112. Weimer PJ. Redundancy, resilience, and host specificity of the ruminal microbiota: implications for engineering improved ruminal fermentations. Front Microbiol 2015; 6:296
https://doi.org/10.3389/fmicb.2015.00296



113. Li F, Li C, Chen Y, et al. Host genetics influence the rumen microbiota and heritable rumen microbial features associate with feed efficiency in cattle. Microbiome 2019; 7:92
https://doi.org/10.1186/s40168-019-0699-1



114. Malmuthuge N, Guan LL. Understanding host-microbial interactions in rumen: searching the best opportunity for microbiota manipulation. J Anim Sci Biotechnol 2017; 8:8
https://doi.org/10.1186/s40104-016-0135-3



- TOOLS
-
METRICS

-
- 1 Crossref
- 0 Scopus
- 1,509 View
- 81 Download
- Related articles






 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print




