 |
 |
| Anim Biosci > Volume 37(4); 2024 > Article |
|
Abstract
Objective
In this study, we aimed to evaluate the usability single nucleotide polymorphisms (SNPs) for parentage testing of horse breeds in Korea.
Methods
The genotypes of 93 horse samples (38 Thoroughbred horses, 17 Jeju horses, 20 Quarter horses, and 18 American miniature horses) were determined using 15 microsatellite (Ms) markers (AHT4, AHT5, ASB2, ASB17, ASB23, CA425, HMS1, HMS2, HMS3, HMS6, HMS7, HTG4, HTG10, LEX3, and VHL20) and 101 SNP markers.
Results
Paternity tests were performed using 15 Ms markers and 101 SNP markers in Thoroughbred horses and Quarter horses. AHT5, ASB2, ASB17, ASB23, CA425, HMS7, HTG10, and LEX3 did not follow Mendelian inheritance in Thoroughbred horses, whereas in Quarter horses, only AHT4, ASB2, and HMS2 showed Mendelian inheritance, consequently, paternity was not established. Meanwhile, 31 markers, including MNEc_2_2_2_98568918_BIEC2_502451, in Thoroughbred horses, and 30 markers, including MNEc_2_30_7430735_BIEC2_816793, in Quarter horses did not conform with Mendelian inheritance and therefore, could not be used for establishing parentage.
The registration of Thoroughbred horses in Korea is implemented in accordance with the International Agreement on Breeding and Racing and the domestic Thoroughbred Horse Registration Regulations. Individual identification and parentage verification for registration of Thoroughbred horses had been conducted using blood-typing tests for nearly 40 years. However, blood-typing tests have the following disadvantages: i) they require fresh whole blood for testing, however, the cost of blood collection by veterinarians and transportation to the laboratory are high; ii) labor costs are high because of the long testing time and the manual nature of the tests; and iii) the accuracy of the results is low, at 97.0% to 99.5%. Because of these reasons, in the early 2000s, microsatellite (Ms) DNA typing was introduced to replace blood-typing tests in Korea for horse parentage testing, and its use has been continued till present day. For Ms testing, semiautomatic tests of blood or hair roots samples can be performed using analytical equipment, with approximately 99.99% accuracy of results [1].
Recent, research in many countries has focused on the introduction of the single nucleotide polymorphism (SNP) method, to replace Ms in horse registration [2,3].
According to the appropriate number and selection of SNP markers, SNPs genotypes can be used for estimating genomic breeding values for multiple traits [4], population monitoring and diversity management [5], and parentage control [6]. Compared with Ms markers, SNPs have a low mutation rate and afford a clear standardization of alleles, as reviewed by Vignal et al [7]. Concomitantly, it facilitates the laboratory automation of the analytical process, which reduces costs and enables a higher throughput in the generation of SNP genotype data [7,8].
Against this background, in this study, SNPs were analyzed and compared with the Ms results with the aim of securing basic data for future horse registration based on SNPs.
This study was a routine test for the registration of Thoroughbred horses in Korea and was not approved by the Research Ethical Committee, however, in this study, sampling was performed according to the guidelines for the care and use of experimental animals of the Animal Ethics Committee of the Korea Racing Authority.
Genomic DNA was obtained from 93 horse hair roots samples (38 Thoroughbred horses, 17 Jeju horses, 20 Quarter horses, and 18 American miniature horses) using an automated extraction system with a Nimbus KingFisher Presto instrument (Thermo Fisher Scientific, Inc, Waltham, MA, USA), according to the manufacturer’s protocols [9]. These horses are four genetically independent breeds (light breed and pony breed). The Thoroughbred horses used in this study for parental judgment were referred to the sample of horses registered in South Korea. For the concentration and purification of the sample, 1.2 μL of the extracted DNA was measured using an Epoch microplate spectrophotometer and the Gen5 3.00 program (Agilent Technology, Santa Clara, CA, USA). Samples that yielded a single band near 23 kb on electrophoresis (0.5× Tris-acetate-ethylenediaminetetraacetic acid, 1% agarose gel, 100 V, 30 min) with a DNA concentration of 10 ng/μL or more and an A260/A280 ration of 1.5 or more were deemed suitable for analysis.
Quality control (QC) and quality assurance of genotype data for genome-wide association studies were performed according to the method reported by Laurie et al [10].
SNP analysis was performed in four stages (DNA amplification, DNA fragmentation, DNA resuspension and hybridization, and ligate-wash image scanning), followed by analysis using the Axiom Equine genotype sequence (Axiom MNEC670; ThermoFisher Scientific GeneTitan MC Instrument, USA).
A total of 15 microsatellite loci (AHT4, AHT5, ASB2, ASB17, ASB23, CA425, HMS1, HMS2, HMS3, HMS6, HMS7, HTG4, HTG10, LEX3, and VHL20) were used for the analysis of the horses. Polymerase chain reaction (PCR) was performed according to the manufacturer’s protocols (Equine Genotypes Panel 1.1 Kit; ThermoFisher Scientific, USA). PCR products were tested using an automatic genetic analyzer (AB 3500XL Genetic Analyzer; ThermoFisher Scientific, USA); subsequent electrophoresis was performed on a POP 7 polymer (ThermoFisher Scientific, USA) at 15 kV. Using peak row data, the size of the alleles (in base pairs) of each marker was determined based on the results of the 2017/2018 Horse Comparison Test No. 1 of the International Society for Animal Genetics (ISAG), using 3500XL Data Collection ver. 5.0 and GeneMapper Software ver. 5.0 (ThermoFisher Scientific, USA). Alleles were showed using alphabetical symbols, in the order of smallest to largest, based on the assignment of a middle-sized allele as M. These definitions were confirmed by the International of Society and Animal Genetics (ISAG) horse comparison test.
The SNP call rate, Fisher’s linear discriminant (FLD), heterozygous strength offset (HetSO), and homozygote ratio offset (HomRO) were analyzed according to the manufacturer’s protocols [11]. The SNP minor allele frequency (MAF) values were analyzed according to the method reported by Plzhnikov et al [12]. A shift in the MAFs can reflect mis-clustering events over the samples on the plates used. Chi-squared analysis is a simple method for automatically detecting this type of effect [12]. The allele frequencies of each SNPs and Ms markers were determined by direct counting from the tests performed for the 93 horses. Based on the SNPs and Ms markers results obtained in this study, we compared parentage testing using SNP and Ms in the same Thoroughbred horses and Quarter horses.
According to the analysis of 101 SNP markers recommended by ISAG in 93 horses, the HomeFLD ranged from 5.600 (MNEc_2_3_87970209_BIEC2_7996664) to 26.199 (MNEc_2_2_78264598_BIEC2_491394), with an average of 15.611. In turn, the HetSO averaged 0.369 and ranged from 0.000 (MNEc_2_3_87970209_BIEC2_799664) to 0.769 (MNEc_2_12_14391372_BIEC2_183251). Moreover, the HomeRO averaged 1.644 and ranged from 0.292 (MNEc_2_5_76618727_BIEC2_919835) to 3.056 (MNEc_2_2_2_78264598_BIEC2_491394). The MAF averaged 0.420 and ranged from 0.101 (MNEc_2_3_87970209_BIEC2_7996664) to 0.500 (MNEc_2_8_91677016_BIEC2_1066179). Finally, the Hardy–Weinberg equilibrium p-value averaged 0.398 and ranged from 0.000 (MNEc_2_4_78695209_BIEC2_870568) to 0.976 (MNEc_2_5_49304134_BIEC2_908630) (Table 1). As shown in Figure 1, FLD (10.09), HetSO (0.56), and HomRO (1.58) indicted a good SNP QC.
The results of the paternity test using Ms markers and SNPs for each foal among Thoroughbred horses and Quarter horses are reported in Tables 2 and 3. Among the 15 Ms markers, AHT5, ASB2, ASB17, ASB23, CA425, HMS7, HTG10, and LEX3 did not obey Mendel’s genetic laws in Thoroughbred horses, whereas in Quarter horses, the AHT4, ASB2, and HMS2 markers alone were consistent with Mendel’s genetic law, therefore, paternity was not established.
In turn, among the 101 SNPs analyzed here, 31 SNPs (including MNEc_2_2_2_98568918_BIEC2_502451) in Thoroughbred horses, and 30 SNPs (including MNEc_2_30_7430735_BIEC2_816793) in Quarter horses were not established for parentage testing because of disobedience of Mendel’s genetic law.
The objective of the present study was to construct a parentage-testing system for parentage testing of horse breeds (including Thoroughbred horses) using SNP markers, and to evaluate its utility for routine parentage testing by comparing it to the system used currently, which is based on Ms markers.
For over two decades, parentage control for horses in Korea has been carried out using the Ms parentage panel recommended by the ISAG (https://www.isag.us/) [13]. In Korea, 15 Ms markers are used to conduct parentage testing for approximately 2,000 Thoroughbred horses each year. Parentage verification is determined using Mendel’s genetic law. If a discrepancy is detected in one marker after the test judgment, it is assumed to be a mutation and additionally examined using three to five TKY (TKY279, TKY301, TKY312, TKY321, TKY341) markers; if two or more markers deviate from Mendel’s genetic law, it is finally established that paternity can be ruled out [1].
Although the use of Ms markers sometimes results in slippage mutation errors, these genotyping systems are widely used by horse registries worldwide. In turn, SNPs are biallelic sequences that are abundantly dispersed throughout most eukaryotic genomes [14–16] and exhibit the following characteristics: i) they have very low mutation rates compared with Ms markers (10 to 8 vs 10 to 3); ii) because of their short length they are highly suitable for analysis using automated high-throughput technologies; and iii) they can be detected using various techniques, such as TaqMan assays [17]. Therefore, there is a growing interest in the application of SNPs in the field of forensics [18,19]. Recently, the interest in the use of SNPs for parentage testing among domestic animals has increased [20,21]; moreover with the completion of the horse genome project, numerous SNPs have also become available for genetic studies in horses. One of the main proposed benefits of SNP-based systems is the potential for genetic diagnosis, such as the determination of coat color [22] and diseases, in horses, which are expected to help the actual coat color information and clinical symptoms gained by observation, because coat color information is particularly used currently for horse identification and registry. Therefore, this is a flexible technology that can be improved to serve further demands; for example, for the genotyping of factors such as coat color. However, there is insufficient information on their utility and related research to SNPs including parentage testing in horses in Korea. This study is the first discovery on horse parentage testing using SNPs; it shows the potential of SNPs testing as a basic tool for improving and developing the current system of horses parentage testing in Korea. This study confirmed that SNPs can be used in place of Ms markers for parentage testing in horses. However, further research using more samples is needed.
Notes
Figure 1
Results of the analysis of single nucleotide polymorphisms (SNPs) QC parameters (FLD, HetSO, and HomRO). (a) Fishers linear discriminant (FLD): measurement of the cluster quality of a SNP. High-quality SNP clusters exhibit: i) well-separated centers, and ii) narrow clusters. Default ≥ 3.6. (b) Heterozygous strength offset (HetSO): measures how far the heterozygous cluster center sits above or below the homozygous cluster centers in the size dimension (y axis). Default ≥ −0.1. (c) Homozygous ratio offset (HomRO): measures the distance to zero in the contrast dimension (x axis) from the center of the homozygous cluster that is closest to zero. The minimum distance is between zero and BB, HomRO = 1.58. Default: ≥−0.9.
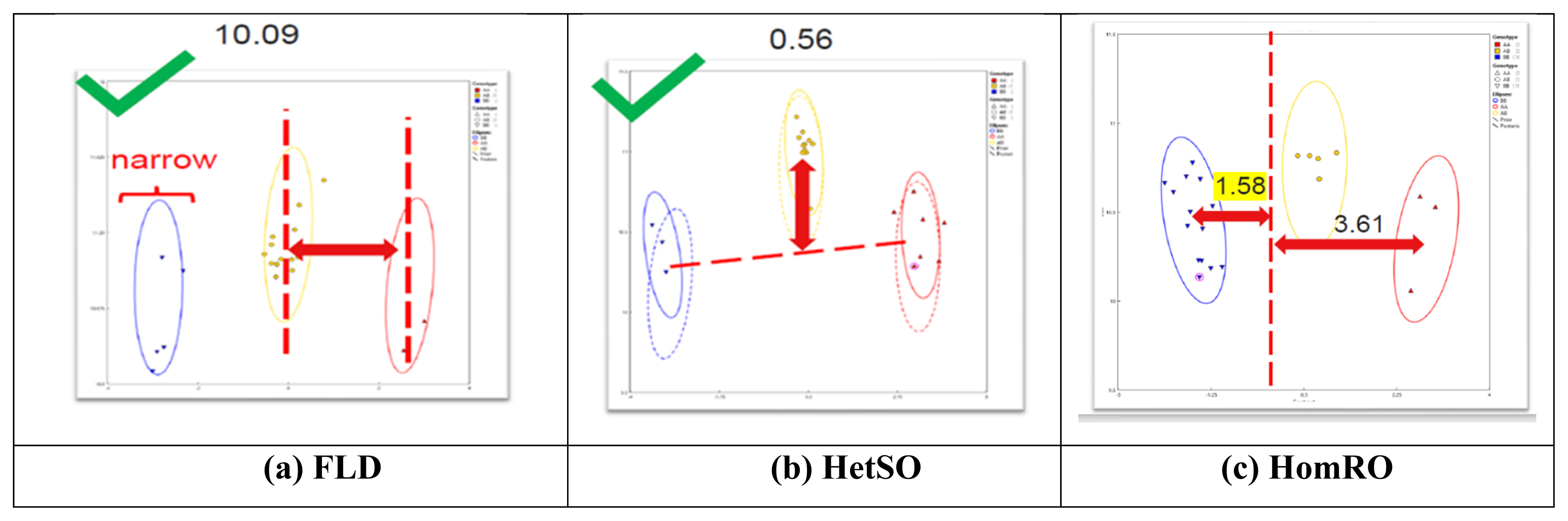
Table 1
HomFLD, HetSO, HOmRO, MAF, H.W. p-value of 101 SNPs in 93 horses
Table 2
A case of exclusion in horse parentage test using microsatellite markers
Table 3
A case of exclusion in horse parentage test using SNPs
REFERENCES
1. Park CS, Lee SY, Cho GJ. Evaluation of recent changes in genetic variability in Thoroughbred horses based on microsatellite markers parentage panel in Korea. Anim Biosci 2022; 35:527–32.
https://doi.org/10.5713/ab.21.0272



2. Hirota KI, Kakoi H, Gawahara H, Hasegawa T, Tozaki T. Construction and validation of parentage testing for thoroughbred horses by 53 single nucleotide polymorphisms. J Vet Med Sci 2010; 72:719–26.
https://doi.org/10.1292/jvms.09-0486


3. Nolte W, Alkhoder H, Wobbe M, et al. Replacement of microsatellite markers by imputed medium density SNP arrays for parentage control in German warmblood horses. J Appl Genet 2022; 63:783–92.
https://doi.org/10.1007/s13353-022-00725-9



4. Meuwissen T, Hayes B, Goddard M. Genomic selection: a paradigm shift in animal breeding. Anim Front 2016; 6:6–14.
https://doi.org/10.2527/af.2016-0002

5. Lee YS, Lee WJ, Kim H. Estimating effective population size of thoroughbred horses using linkage disequilibrium and theta (4Nμ) value. Livest Sci 2014; 168:32–7.
https://doi.org/10.1016/j.livsci.2014.08.008

6. Holl HM, Vanhnasy J, Everts RE, et al. Single nucleotide polymorphisms for DNA typing in the domestic horse. Anim Genet 2017; 48:669–76.
https://doi.org/10.1111/age.12608


7. Vignal A, Milan D, SanCristobal M, Eggen A. A review on SNP and other types of molecular markers and their use in animal genetics. Genet Sel Evol 2002; 34:275
https://doi.org/10.1186/1297-9686-34-3-275



8. McClure MC, Sonstegard TS, Wiggans G, et al. Imputation of microsatellite alleles from dense SNP genotypes for parentage verification across multiple Bos taurus and Bos indicus breeds. Front Genet 2013; 4:176
https://doi.org/10.3389/fgene.2013.00176



9. Thermo Fisher Scientific. KingFisher Presto User Manual 2016.
10. Laurie CC, Doheny KF, Mirel DB, et al. Quality control and quality assurance in genotypic data for genome-wide association studies. Genet Epidemiol 2010; 34:591–602.
https://doi.org/10.1002/gepi.20516



11. Thermo Fisher Scientific. Axiom™ Genotyping Solution Data Analysis User Guide. Santa Clara, CA, USA: Thermo Fisher Scientific Inc; 2022.
12. Pluzhnikov A, Below JE, Tikhomirov A, Konkashbaev A, Nicolae D, Cox NJ. Differential bias in genotype calls between plates due to the effect of a small number of lower DNA quality and/or contaminated samples. Genet Epidemiol 2008; 32:676
13. Bowling AT, Eggleston-Stott ML, Byrns G, Clark RS, Dileanis S, Wictum E. Validation of microsatellite markers for routine horse parentage testing. Anim Genet 1997; 28:247–52.
https://doi.org/10.1111/j.1365-2052.1997.00123.x


14. McKay SD, Schnabel RD, Murdoch BM, et al. An assessment of population structure in eight breeds of cattle using a whole genome SNP panel. BMC Genet 2008; 9:37
https://doi.org/10.1186/1471-2156-9-37



15. Petkov PM, Ding Y, Cassell MA, et al. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res 2004; 14:1806–11.
https://doi.org/10.1101/gr.2825804



16. Salisbury BA, Pungliya M, Choi JY, Jiang R, Sun XJ, Stephens JC. SNP and haplotype variation in the human genome. Mutat Res 2003; 526:53–61.
https://doi.org/10.1016/s0027-5107(03)00014-9


17. Hui L, DelMonte T, Ranade K. Genotyping using the TaqMan assay. Curr Protoc Hum Genet 2008; 56:2.10.1–8.
https://doi.org/10.1002/0471142905.hg0210s56

18. Lee HY, Park MJ, Yoo JE, Chung U, Han GR, Shin KJ. Selection of twenty-four highly informative SNP markers for human identification and paternity analysis in Koreans. Forensic Sci Int 2005; 148:107–12.
https://doi.org/10.1016/j.forsciint.2004.04.073


19. Lessig R, Zoledziewska M, Fahr K, et al. Y-SNP genotyping—a new approach in forensic analysis. Forensic Sci Int 2005; 154:128–36.
https://doi.org/10.1016/j.forsciint.2004.09.129


20. Rohrer GA, Freking BA, Nonneman D. Single nucleotide polymorphisms for pig identification and parentage exclusion. Anim Genet 2007; 38:253–8.
https://doi.org/10.1111/j.1365-2052.2007.01593.x


21. Werner FAO, Durstewitz G, Habermann FA, et al. Detection and characterization of SNPs useful for identity control and parentage testing in major European dairy breeds. Anim Genet 2004; 35:44–9.
https://doi.org/10.1046/j.1365-2052.2003.01071.x


22. Haase B, Brooks SA, Tozaki T, et al. Seven novel KIT mutations in horses with white coat colour phenotypes. Anim Genet 2009; 40:623–9.
https://doi.org/10.1111/j.1365-2052.2009.01893.x








 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print





