 |
 |
Abstract
Four Korean native cattle (KNC) breedsŌĆöHanwoo, Chikso, Heugu, and Jeju blackŌĆöare entered in the Domestic Animal Diversity Information System of the United Nations Food and Agriculture Organization (FAO). The objective of this study was to assess the genetic diversity, phylogenetic relationships and population structure of these KNC breeds (n = 120) and exotic breeds (Holstein and Charolais, n = 56). Thirty microsatellite loci recommended by the International Society for Animal Genetics/FAO were genotyped. These genotypes were used to determine the allele frequencies, allelic richness, heterozygosity and polymorphism information content per locus and breed. Genetic diversity was lower in Heugu and Jeju black breeds. Phylogenetic analysis, Factorial Correspondence Analysis and genetic clustering grouped each breed in its own cluster, which supported the genetic uniqueness of the KNC breeds. These results will be useful for conservation and management of KNC breeds as animal genetic resources.
Korean cattle have existed in the Korean Peninsula for at least 2000 years (Kim and Lee, 2000). A mural in a tomb dating from the Kokuryo Age (ŌĆ£Anak-3-hobunŌĆØ, A.D. 357) depicts three cattle heads that differ in coat color (brown, brindle and black) (Na, 2008). These ancient animals are thought to be the origin of the native cattle breeds existing in Korea. Four Korean native cattle (KNC) breeds have been recently documented in the Domestic Animal Diversity Information System of the United Nations Food and Agriculture Organization (FAO; http://dad.fao.org/)ŌĆö Hanwoo (HW), Chikso (CS), Heugu (HU), and Jeju black (JB). These breeds are classified based on different coat colors (HU and JB, black; HW, brown; CS, brindle) (Figure 1) and geographical distribution (HW is widely distributed in Korea, JB only on Jeju Island and HU and CS on the Korean peninsula, except Jeju Island).
Beginning in the 1960ŌĆÖs, the Korean government embarked on a strategy aimed at enhancing the performance and genetic ability of HW. The program was successful and now HW is one of the superior commercial livestock breeds in Korea. The other breeds had been except from this governmental plan and their existence became precarious (MAF, 2004). In response the government committed to the conservation and proliferation of JB, HU, and CS, given their recognition as valuable genetic resources for development of new beef cattle different from HW.
High polymorphic nuclear markers are now commonly used in the evaluation of genetic diversity, phylogenetic relationship and population structure within and among livestock breeds (Groeneveld et al., 2010). Microsatellite analyses have provided useful genetic information for European (Maudet et al., 2002; Wiener et al., 2004; Padilla et al., 2009), African (Dadi et al., 2008), mid-South American (Egito et al., 2007; Acosta et al., 2013) and Asian cattle breeds (Zhang et al., 2007; Pham et al., 2013). In Korea, several studies have evaluated the genetic diversity and genetic relationship of HW compared with other breeds (Kim et al., 2002; Choi et al., 2012). However, little genetic characterization of JB, HU, and CS has been done.
The aims of the present study were to evaluate the extent of the genetic diversity of KNC breeds, and to establish the relationships between the four Korean native and exotic cattle breeds using 30 microsatellite markers.
Blood from 30 CS individuals were sampled from two local institutes (Gangwon Provincial Livestock Research Center and Chungbuk Veterinary Service Center). Blood was collected from HU (n = 30) and JB (n = 30) from Chungbuk Veterinary Service Center and Jeju Special Self-Governing Provincial Livestock Institute, respectively. Blood samples of these three breeds were randomly collected, while avoiding parent-offspring or sib pairs where possible according to pedigree information of each institute. The HW (n = 30) was additionally sampled for blood from 11 farms (1 to 7) in Yeongju City.
Genomic DNA from blood samples was extracted using the DNeasy Blood Kit (Qiagen, Germany) following the manufacturerŌĆÖs procedure. Genomic DNA of two exotic breeds, Holstein (n = 30) and Charolais (n = 26) was obtained from the Animal Genetic Resources Station, National Institute of Animal Science, Rural Development Administration.
Thirty microsatellite markers (BM1818, BM1824, BM2113, CSRM60, CSSM66, ETH3, ETH10, ETH152, ETH185, ETH225, HAUT24, HAUT27, HEL1, HEL5, HEL9, HEL13, ILSTS005, ILSTS006, INRA005, INRA023, INRA032, INRA035, INRA037, INRA063, MM12, SPS115, TGLA53, TGLA122, TGLA126, and TGLA227) were analyzed to estimate various parameters of genetic diversity. Microsatellites were amplified in multiplexes (3 to 4 co-amplified loci), with the exception of ETH185. Polymerase chain reaction amplification was performed in a 15 ╬╝L reaction mixture, which contained 1.5 mM MgCl2, 1 U of AmpliTaq Gold (Applied Biosystems, Foster, CA, USA), 3ŌĆō10 pmol of each forward (labeled with a fluorescent-colored dye) and reverse primer and approximately 10 ng genomic DNA as a template.
The PCR amplification comprised an initial denaturation at 95┬░C for 15 min, followed by 35 cycles of denaturation at 95┬░C for 30 s, annealing at 55┬░C to 63┬░C (respective optimal annealing temperature) for 90 s, extension of starters at 72┬░C for 90 s and a final extension of starters at 72┬░C for 40 min, using the GeneAmp PCR System 9700 (Applied Biosystems). Electrophoresis was carried out using an ABI 3130xl Genetic Analyzer (Applied Biosystems). Allele sizes of each microsatellite were determined using GeneMapper ver. 4.0 (Applied Biosystems). The formulated allele data was used for statistical analyses.
Cervus ver. 3.0.3 (Kalinowski et al., 2007) was used to estimate allele frequencies, total number of alleles (TNA), mean observed (HObs) and expected (HExp) heterozygosities ad mean polymorphism information content (PIC) per locus and breed. Allelic richness (AR) for each breed was calculated to correct distortion by sample size difference using FSTAT ver. 2.9.3.2. (Goudet, 2002). The DA genetic distance (Nei et al., 1983) was calculated with MSA (Dieringer and Schl├Čtterer, 2003). The three dimensions of a multivariate factorial correspondence analysis (FCA) were computed using GENETIX ver. 4.05 (Belkhir et al., 2004) from allele doses for each individual. The FCA was carried out from all animals and for the 30 loci. Genetic structure and the degree of admixture of KNC breeds were investigated using the Bayesian clustering procedure of STRUCTURE ver. 2.3 (Pritchard et al., 2000). Fifty independent runs were performed for each K between 2 and 10, with a burn-in period of 50,000 iterations followed by 100,000 iterations of the Markov chain Monte Carlo algorithm. To identify the most probable groups (K) that best fit the data, we used the STRUCTURE HARVESTER (Earl and von Holdt, 2012), which implements the Evanno method (Evanno et al., 2005). The program CLUMPP ver. 1.1 (Jakobsson and Rosenberg, 2007) was used to align the 50 repetitions of each K. The CLUMPP out files were visualized using DISTRUCT ver. 1.1 (Rosenberg, 2004).
Allele ranges, number of alleles, heterozygosity and PIC per locus are summarized in Table 1. A total of 276 alleles were detected at 30 microsatellite loci across four KNC breeds. The TNA per locus ranged from 4 (ILSTS005) to 17 (TGLA122), with a mean of 9.20┬▒0.58 alleles. The mean of HExp across loci was 0.733┬▒0.018, with estimates per locus ranging from 0.473 (ILSTS005) to 0.893 (TGLA53). For HObs, the mean for all loci was 0.667┬▒0.028, and the range was between 0.174 (INRA035) and 0.855 (CSRM60).
The 30 microsatellite markers used were recommended from by International Society for Animal Genetics/FAO (Hoffmann et al., 2004). Most of the loci were highly informative (PIC>0.5), with the exception of ILSTS005 (0.375) and HEL13 (0.413). Similarly, ILSTS005 and HEL13 have been reported to be the relatively low informative markers (Padilla et al., 2009). According to Botstein et al. (1980), PIC of >0.5 indicates a highly informative locus for chromosomal mapping and genetic diversity. Therefore, most microsatellite marker sets are highly informative and useful for evaluation of genetic diversity and population structure in KNC breeds.
The various indices of genetic diversity across the four KNC breeds are shown in Table 2. The mean number of alleles and AR ranged from 4.73┬▒0.32 (HU) to 6.97┬▒0.41 (HW) and from 4.39┬▒0.28 (HU) to 6.42┬▒0.37 (HW), respectively. The mean value of HExp was highest in HW (0.713) and lowest in JB (0.604). The mean value of HObs ranged from 0.613 (HU) to 0.683 (CS). Generally, the levels of genetic diversity of HW and CS breeds were higher than those of HU and JB. The difference of the levels of genetic diversity among Korean cattle breeds could be explained by population sizes and distribution region. Populations of HW and CS are relatively large (currently 3,000,000 animals) and medium-sized (currently 1,700 animals), respectively. These two breeds are widely distributed throughout Korea. On the other hand, HU and JB are numerically small populations, with 300 to 400 animals each. Moreover, JB are restricted in their distribution to Jeju Island. HU and JB are considered endangered breeds (IUCN, 2000). The HExp and HObs values observed in KNC breeds are reportedly similar or higher than those of British and Spanish cattle breeds (HExp = 0.56 to 0.68 and HObs = 0.59 to 0.67) (Wiener et al., 2004; Martin-Burriel et al., 2007; Padilla et al., 2009). The mean values of HObs in HW and CS breeds were lower than those of HExp. Generally, the mating of the four KNC breeds has been non-randomly performed by using limited bulls. In addition, the number of bulls used is fewer in HW and CS than in HU and JB based on population size.
To verify the genetic relationship among the four Korean native and two exotic cattle breeds, we calculated the DA distance and constructed a phylogenetic tree. Among the Korean native breeds, HW and CS were closest (DA = 0.129), with the largest difference observed for JJ and HU (DA = 0.265) (Table 3). The phylogenetic tree was constructed from a neighbor joining (NJ) clustering based on the DA distance matrix among breeds (Figure 2). The genetic relationship between HW and CS was relatively close among the Korean breeds, whereas HU and JJ were distinctly separated. The NJ tree indicated a clear separation of two exotic breeds (HT, CR) from the four KNC breeds. Yoon et al. (2005) emphasized that genetic similarity was high among Hanwoo, Heugu, two Japanese breeds, and a Chinese breed (Yanbian). Additionally, the bootstrap values among these breeds ranged from 40 to 50. Thus, the low values in this study were caused by high genetic similarity among Korean native breeds.
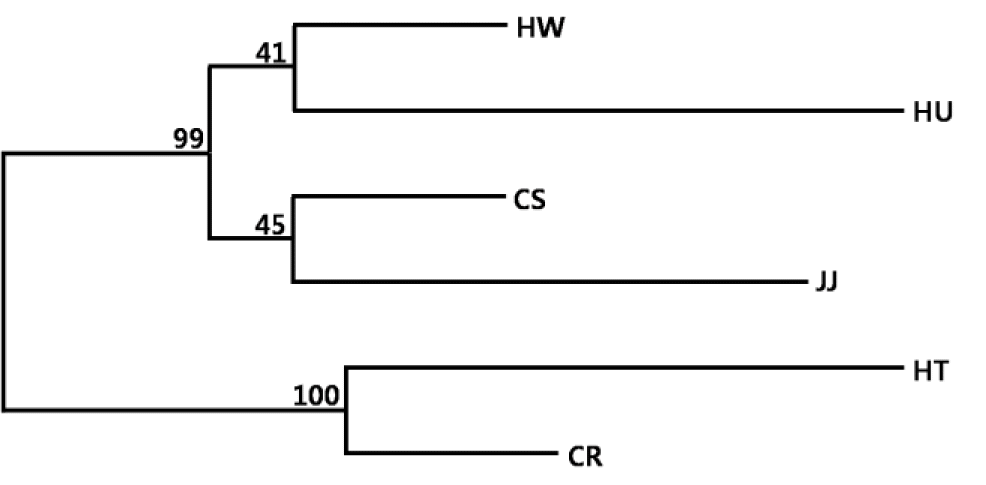
The FCA analysis revealed the very clear separation between the HT, CR, HU, JJ, and other two Korean breeds (HW and CS) (Figure 3). About 75% of the variance was accounted for by the first to three dimensions of the FCA Axis 1 (33.33% of total variance explained), which separated HT and CR from the Korean native breeds. Axis 2 (24.37%) further separated HU, JJ and the HW and CS Korean breeds. Axis 3 (17.20%) distinctly separated HT and CR. These results indicated that individuals from HW and CS were mixed, suggesting a closer relationship between them. DA distance, phylogenetic tree and FCA results provide genetic evidence for the differentiation of the four Korean cattle breeds.
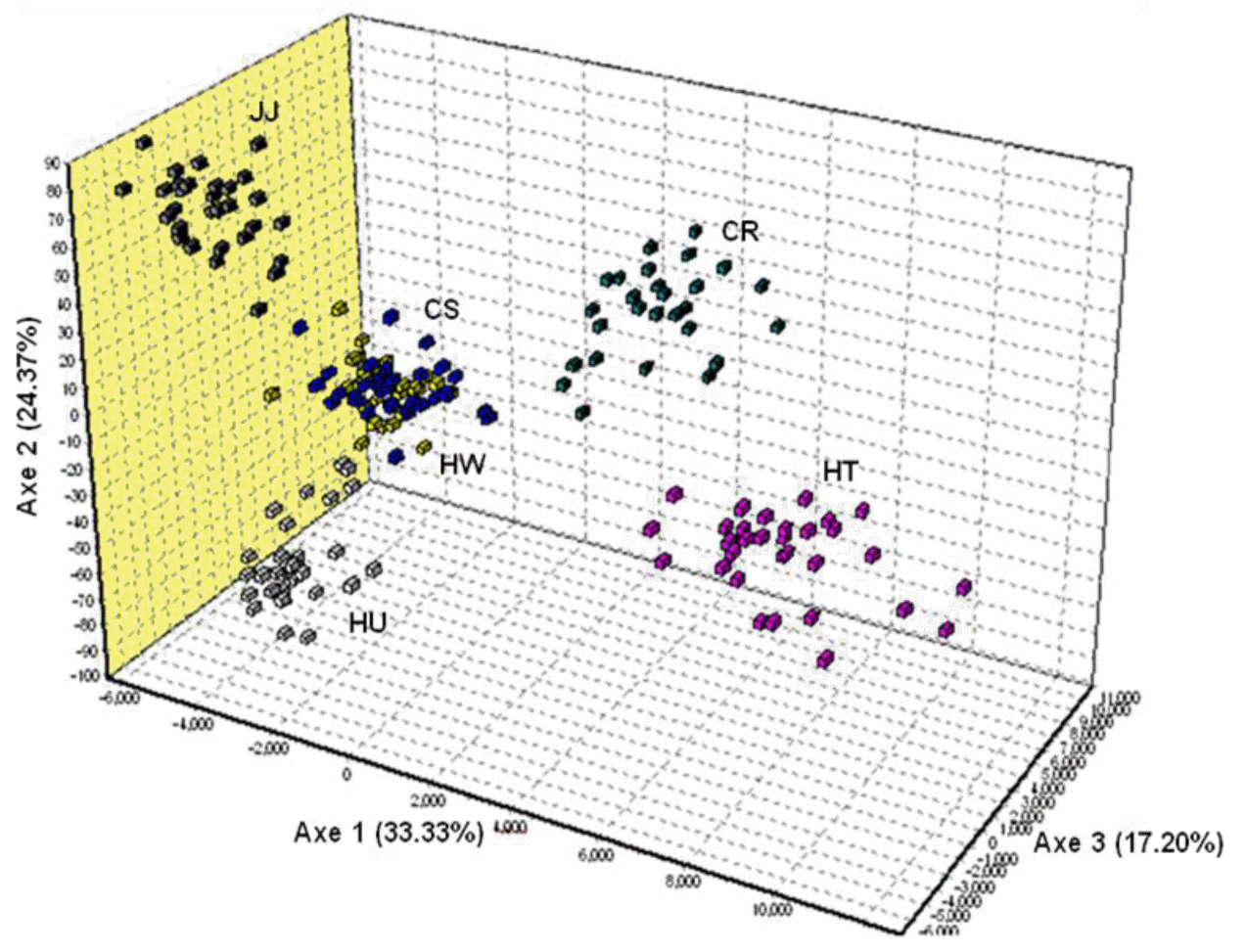
STRUCTURE software was used to determine the unbiased structure without prior knowledge regarding the number of breeds (Figure 4). At K = 2, two main groups that accurately corresponded to Korean native and exotic cattle breeds were formed. As K increased, the contributions of the assumed populations resulted in the progressively complete separation of the 6 breeds. The largest delta K (ΔK) value was calculated as previously described (Evanno et al., 2005). The optimum ΔK value (ΔK = 92.94, data not shown) was found at K = 5. These results failed to differentiate between the HW and CS breeds. However, each breed grouped in its own cluster with an estimated membership >0.893 at K = 6 (ΔK = 52.54) (Figure 4, Table 4). According to Leroy et al. (2009), the highest ΔK values can potentially reveal the optimal K, but some weakly defined substructures can be found when only a small number or breeds are analyzed. In addition, Kim et al. (2013) suggested that CS and HW have a genetic difference based on sequence variation and phylogenetic analysis of mtDNA cyt b gene. Based on these reports, we considered that HW and CS could have distinct genetic characteristics.
In this study, the level of genetic diversity among KNC breeds differed according to their population sizes (endangered, vulnerable or not at risk). However, these were higher than those of several native cattle breeds of other countries. In analyses of genetic relationship and clustering, all KNC breeds were genetically differentiated from the two exotic breeds. In addition, the scientific evidence supports the genetic differentiation among four KNC breeds. The results suggest that each KNC breed had distinct breed-specific genetic characteristics. The results of this study may be useful as scientific evidence to design plans for future conservation, improvement and breed management of each KNC breed.
ACKNOWLEDGMENTS
This work was carried out with the support of ŌĆ£Cooperative Research Program for Agriculture Science and Technology Development (Project title: Molecular genetic monitoring of conservation population of animal genetic resources, Project No. PJ008431)ŌĆØ Rural Development Administration, Republic of Korea. This study was supported by 2014 Postdoctoral Fellowship Program of National Institute of Animal Science, Rural Development Administration, Republic of Korea.
Figure┬Ā1
Different coat color phenotypes of the Korean native cattle breed; (a) Hanwoo, (b) Chikso, (c) Heugu, (d) Jeju black.

Figure┬Ā2
Phylogenetic tree constructed from DA by the neighbor-joining method showing the genetic relationships among six cattle breeds. The values at the nodes are the percentages of bootstrap values from 1,000 replications of re-samplings. HW, Hanwoo; CS, Chikso; HU, Heugu; JJ, Jeju black; HT, Holstein; CR, Charolais.
Figure┬Ā3
Factorial Correspondence Analysis of individual cattle microsatellite genotypes calculated using GENETIX. HW, Hanwoo; CS, Chikso; HU, Heugu; JJ, Jeju black; HT, Holstein; CR, Charolais.
Figure┬Ā4
Population structure of the analyzed six cattle breeds without prior population affiliation using a model-based clustering method implemented in STRUCTURE for K = 2 to K = 6. Each column represents the proportion in which an individual belongs to a different coloured cluster. HW, Hanwoo; CS, Chikso; HU, Heugu; JJ, Jeju black; HT, Holstein; CR, Charolais.
Table┬Ā1
Polymorphism of 30 microsatellite loci across the four Korean cattle breeds
Table┬Ā2
Genetic diversity parameters estimated for 30 microsatellite loci in four Korean native cattle breeds
N, number of animals; TNA, total number of alleles with standard error; MNA, mean number of alleles with standard error; AR, allelic richness with standard error; HExp, expected heterozygosity with standard error; HObs, observed heterozygosity with standard error; PIC, polymorphism information content with standard error.
Table┬Ā3
NeiŌĆÖs genetic distance (DA) values among six cattle breeds. Maximum and minimum values are shown in bold
| Breed | ||||||
|---|---|---|---|---|---|---|
|
|
||||||
| HW | CS | HU | JJ | HT | CR | |
| HW | - | |||||
| CS | 0.129 | - | ||||
| HU | 0.185 | 0.193 | - | |||
| JJ | 0.181 | 0.180 | 0.265 | - | ||
| HT | 0.239 | 0.227 | 0.311 | 0.316 | - | |
| CR | 0.194 | 0.193 | 0.274 | 0.255 | 0.188 | - |
Table┬Ā4
Population of membership of each the six cattle population genotypes with the ISAG/FAO recommended 30 microsatellite markers in the six inferred clusters using STRUCTURE analysis
REFERENCES
Acosta AC, Uffo O, Sanz A, Ronda R, Osta R, Rodellar C, Martin-Burriel I, Zaragoza P. 2013. Genetic diversity and differentiation of five Cuban cattle breeds using 30 microsatellite loci. J Anim Breed Genet 130:79ŌĆō86.


Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. 2004. GENETIX ver. 4.05, Logiciel sous WindowsTM pour la G├®n├®tique des Populations. Laboratoire G├®nome et Population, Universit├® Montpellier II; Montpellier, France: http://kimura.univ-montp2.fr/genetix/Accessed May 30, 2014
Botstein D, White RL, Skolnick M, Davis RW. 1980. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314ŌĆō331.


Choi TJ, Lee SS, Yoon DH, Kang HS, Kim CD, Hwang IH, Kim CY, Jin X, Yang CG, Seo KS. 2012. Determination of genetic diversity among Korean Hanwoo cattle based on physical characteristics. Asian Australas J Anim Sci 25:1205ŌĆō1215.



Dadi H, Tibbo M, Takahashi Y, Nomura K, Hanada H, Amano T. 2008. Microsatellite analysis reveals high genetic diversity but low genetic structure in Ethiopian indigenous cattle populations. Anim Genet 39:425ŌĆō431.


Dieringer D, Schl├Čtterer C. 2003. Microsatellite analyzer (MSA): A platform independent analysis tool for large microsatellite data sets. Mol. Ecol Notes 3:167ŌĆō169.

Earl DA, vonHoldt BM. 2012. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359ŌĆō361.

Egito AA, Paiva SR, Maria do Socorro , Albuquerque M, Mariante AS, Almeida LD, Castro SR, Grattapaglia D. 2007. Microsatellite based genetic diversity and relationships among ten Creole and commercial cattle breeds raised in Brazil. BMC Genet 8:83



Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol Ecol 14:2611ŌĆō2620.


Goudet J. 2002. FSTAT version 2.9.3.2. department of ecology and evolution. University of Lausanne; Swizeland:
Groeneveld LF, Lenstra JA, Eding H, Toro MA, Scherf B, Pilling D, Negrini R, Finlay EK, Jianlin H, Groeneveld E, Weigend S. 2010. Genetic diversity in farm animalsŌĆōA review. Anim Genet 41:6ŌĆō31.


Hoffmann I, Marsan PA, Barker JSF, Cothran EG, Hanotte O, Lenstra JA, Milan D, Weigend S, Simianer H. 2004. New MoDAD marker sets to be used in diversity studies for the major farm animal species: recommendations of a joint ISAG/FAO working group. In : Proceedings of the 29th International Conference on Animal Genetics; Toyko, Japan. p. 11ŌĆō16.
IUCN (International Union for Conservation of Nature). 2000. The IUCN Red List Categories and Criteria Version 3.1. Gland; Switzerland:
Jakobsson M, Rosenberg NA. 2007. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801ŌĆō1806.


Kalinowski ST, Taper ML, Marshall TC. 2007. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16:1099ŌĆō1106.


Kim JB, Lee C. 2000. Historical look at the genetic improvement in Korean cattle - Review -. Asian Australas J Anim Sci 13:1467ŌĆō1481.

Kim JH, Byun MJ, Kim MJ, Suh SW, Ko YG, Lee CW, Jung KS, Kim ES, Yu DJ, Kim WY, Choi SB. 2013. mtDNA diversity and phylogenetic state of Korean cattle breed, Chikso. Asian Australas J Anim Sci 26:163ŌĆō170.



Kim KS, Yeo JS, Choi CB. 2002. Genetic diversity of north-east Asian cattle based on microsatellite data. Anim Genet 33:201ŌĆō204.


Leroy G, Verrier E, Meriaux JC, Rognon X. 2009. Genetic diversity of dog breeds: between-breed diversity, breed assignation and conservation approaches. Anim Genet 40:333ŌĆō343.


MAF (Ministry of Agriculture and Forestry, Republic of Korea). 2004. National Report on the State of Animal Genetic Resources. Seoul: Rep of Korea; 20ŌĆō21. ftp://ftp.fao.org/docrep/fao/010/a1250e/annexes/CountryReports/KoreanRepublic.pdf/Accessed May 30, 2014
Martin-Burriel I, Rodellar C, Lenstra JA, Sanz A, Cons C, Osta R, Reta M, Arg├╝ello SD, Sanz S, Zaragoza P. 2007. Genetic diversity and relationships of endangered Spanish cattle breeds. J Hered 98:687ŌĆō691.


Maudet C, Luikart G, Taberlet P. 2002. Genetic diversity and assignment tests among seven French cattle breeds based on microsatellite DNA analysis. J Anim Sci 80:942ŌĆō950.


Na GJ. 2008. Characteristics of Korean native cattle (in Korean). Korea Animal Improvement Association Bulletin 1:42ŌĆō52.
Nei M, Tajima F, Tateno Y. 1983. Accuracy of estimated phylogenetic trees from molecular data. J Mol Evol 19:153ŌĆō170.


Padilla J├ü, Sansinforiano E, Parejo JC, Rabasco A, Mart├Łnez-Tranc├│ M. 2009Inference of admixture in the endangered Blanca Cacere├▒a bovine breed by microsatellite analyses. Livest Sci 122:314ŌĆō322.

Pham LD, Do DN, Binh NT, Nam LQ, Ba NV, Thuy TTT, Hoan TX, Cuong VC, Kadarmideen HN. 2013. Assessment of genetic diversity and population structure of Vietnamese indigenous cattle populations by microsatellites. Livest Sci 155:17ŌĆō22.

Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945ŌĆō959.




Rosenberg NA. 2004. DISTRUCT: A program for the graphical display of population structure. Mol. Ecol Notes 4:137ŌĆō138.

Wiener P, Burton D, Williams JL. 2004. Breed relationships and definition in British cattle: A genetic analysis. Heredity 93:597ŌĆō602.


- TOOLS
-
METRICS

- Related articles
-
Genetic diversity of Indonesian cattle breeds based on microsatellite markers2019 April;32(4)





 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print




