 |
 |
Abstract
The objective of this study was to determine the molecular characteristics of the horse vascular endothelial growth factor alpha gene (VEGFα) by constructing a phylogenetic tree, and to investigate gene expression profiles in tissues and blood leukocytes after exercise for development of suitable biomarkers. Using published amino acid sequences of other vertebrate species (human, chimpanzee, mouse, rat, cow, pig, chicken and dog), we constructed a phylogenetic tree which showed that equine VEGFα belonged to the same clade of the pig VEGFα. Analysis for synonymous (Ks) and non-synonymous substitution ratios (Ka) revealed that the horse VEGFα underwent positive selection. RNA was extracted from blood samples before and after exercise and different tissue samples of three horses. Expression analyses using reverse transcription-polymerase chain reaction (RT-PCR) and quantitative-polymerase chain reaction (qPCR) showed ubiquitous expression of VEGFα mRNA in skeletal muscle, kidney, thyroid, lung, appendix, colon, spinal cord, and heart tissues. Analysis of differential expression of VEGFα gene in blood leukocytes after exercise indicated a unimodal pattern. These results will be useful in developing biomarkers that can predict the recovery capacity of racing horses.
Racing horses, such as Thoroughbreds, have been selected for athletic performance traits by implementing systemic structural and functional adaptations. These processes have given racing horses a range of extreme physiological characteristics, including high aerobic and anaerobic metabolic capacities, a large lung volume, high maximum hemoglobin concentration and cardiac output as well as a large muscle mass to body ratio, high skeletal muscle mitochondrial density and oxidative enzyme activity, and large stores of energy substrates (Essén-Gustavsson and Lindholm, 1985; Pösö et al., 1993; Hyyppä, et al., 1997; Hinchcliff and Geor, 2008). Racing, as a sort of exercise, results in acute immune responses and inflammation in joints and muscle of horses (Auer et al., 1989; Bertone et al., 2001; Petersen et al., 2004; Firth et al., 2006). In horses, like human, intense physical activity induces subclinical tissue damage in muscle and joints, resulting in recruit of immune cells, i.e., neutrophils and monocytes. These cells trigger inflammation by up-regulating various cytokines, such as tumor necrosis factor alpha (TNFα), interleukin-1 beta (IL-1β), interleukin-6 (IL-6), and interleukin-10 (IL-10) (Streltsova et al., 2006; Lamprecht et al., 2008) and growth factors such as VEGFα in muscle (Breen et al., 1996; Roca et al., 1998; Richardson et al., 1999; Richardson et al., 2000; Brutsaert et al., 2002).
It is crucial for racing horses to recover from exercise as soon as possible. This recovery process might be mediated by soluble factors such as cytokines and growth factors. Nonetheless, there is no biomarker based on cytokines and/or growth factors that may reflect the degree of recovery from exercise in racing horses. In a previous study, we identified differentially expressed genes (DEG) in muscle and blood from Thoroughbred horses in response to exercise (Park et al., 2012). Furthermore, the findings provides a chance to develop molecular markers that may be related to economic traits of racing horses. The identified DEGs can be used to develop biomarkers that might be useful for monitoring the health of a horse after racing. We found that VEGFα expression was significantly increased in muscle, indicating that VEGFα may play an important role in recovery from exercise. In spite of the importance of VEGFα in various biological processes, such as angiogenesis, so far few studies have been conducted on this gene at the molecular level in horses. Therefore, in this study, we investigated the molecular structure of horse VEGFα gene, constructed a phylogenetic tree, and examined the expression profiles in various horse tissue samples. In addition, since there is a lack of suitable biomarkers to monitor recovery in horses after exercise, we explored the possibility of using VEGFα as a biomarker for exercise and recovery in blood leukocytes.
Blood samples were obtained from three male Thoroughbred horses (2 through 4 year-old; two horses: 2 year-old, one horse: 4 year-old) maintained at Ham-An racing Horse Resort and Training Center. Horses were placed on a treadmill and the treadmill exercises were performed at the speed of 10 to 15 km/h. The blood samples were taken before exercise and every 30 min after exercise up to 120 min. The National Institute of Subtropical Agriculture, Rural Development Administration, provided three male Jeju horses (4 year-old) that were used for tissue sampling. Skeletal muscle, kidney, thyroid, lung, appendix, colon, spinal cord and heart tissues were obtained from each horse and kept in liquid N2 until RNA extraction.
The VEGFα DNA and amino acid sequences of ten species (cattle, chicken, chimpanzee, dog, horse, human, pig, mouse, rat, sheep) were retrieved from Ensembl (http://www.ensembl.org/), The Ensembl ID for the sequences are as follows: horse- ENSECAG00000009402, human- ENSG00000112715, chimpanzee- ENSPTRG000 00001080, mouse-ENSMUSG00000023951, rat- ENS RNOG00000019598, cow- ENSBTAG00000005339, pig: ENSSSCG00000001695, chicken- ENSGALG00000010290, dog- ENSCAFG00000001938, cat- ENSFCAG00000026462.
The retrieved sequences were aligned in BioEdit with the CLUSTERW option. Phylogenetic analysis was conducted using MEGA version 5.0 (Arizona State University, AZ, USA) (Tamura et al., 2011) The Neighbor Joining method (Saitou et al., 1987) was used with the following options: pairwise-deletion, 1,000 bootstrap replications and Kimura 2-parameter. Pairwise deletion was chosen to retain all sites initially, then excluding them as necessary in the pairwise distance estimation. Substitutions of nucleotides were obtained by using the Kimura 2-parameter model. We also showed that synonymous substitutions per site (Ks) and non-synonymous substitutions per site (Ka) in VEGFα genes for individual species. In addition, pairwise distance was calculated by analyzing the nucleotide and amino acid similarity.
Trizol reagent (Invitrogen, Carlsbad, CA, USA) was used to extract total RNA from the tissue samples (skeletal muscle, kidney, thyroid, lung, appendix, colon, spinal cord, and heart) and leukocytes after exercise according to the Invitrogen manual. In order to prevent contamination of genomic DNA, RNase-free DNase kit (Qiagen) was used according to the manufacturer’s manual. Total RNA quantification was performed using a NanoDrop ND-1000 Spectrophotometer (Thermo Scientific, Wilmington, DE, USA). The cDNAs were synthesized in a reaction with oligo-dT primers, moloney-murine leukemia virus (MMLV) reverse transcriptase (Promega, Madison, WI, USA), RNase inhibitor (Promega) and RNase-free ddH2O, which was incubated at 37°C for 4 h.
The horse VEGFα expression level was analyzed by reverse transcription-polymerase chain reaction (RT-PCR) amplification. The primers were designed using the PRIMER3 software (MIT, Cambridge, MA, USA, http://frodo.wi.mit.edu/primer3/) with an expected product size of 184 bp. To detect equine VEGFα mRNA, following primers were used: forward 5′-CTA CCT CCA CCA TGC CAA GT-3′ and reverse 5′-CAC ACA GGA TGG CTT GAA GA-3′. The RT-PCR conditions were as follows: an initial step of 94°C for 10 min, 35 cycles of 94°C for 30 s, 60°C for 30 s, 72°C for 30 s, and a final step of 72°C for 10 min. RT-PCR products were analyzed with gel electrophoresis on a 2.0% SeaKem LE agarose gel (Lonza, NJ, USA). Equine GAPDH gene was used as a normalizer for RT-PCR.
The cDNA was analyzed by quantitative-PCR (qPCR) on BioRad CFX-96 (Bio-Rad, Hercules, CA, USA). Each reaction was performed in a total volume of 25μL with 14 μL SYBR green master mix, 2 μL (5 pmol) VEGFα forward primer, 2 μL (5 pmol) reverse primer, 5 μL distilled water and 2μL (50 ng/μL) of the cDNA. The PCR conditions were as follows: pre-denaturation step at 94°C for 3 min, 39 cycles at 94°C for 10 s and at 60°C for 30 s, 72°C for 30 s, followed by 72°C for 10 min as a final step. Dissociation was accomplished in a condition at temperature increase from 55°C to 95°C over 25 min. Dissociation temperature increased in 0.5°C each. All samples were measured in triplicate to ensure reproducibility, and Ct value was calculated using the 2−ΔΔCt method (Livak et al., 2001). Statistical significance was calculated by t-test. A p values of less than 0.05 were considered to indicate statistical significance.
In a previous study, we performed RNA-seq analyses to identify DEGs in response to exercise. Among the identified DEGs, we found that the transcript of VEGFα was expressed up to 2.59 folds higher after exercise (Park et al., 2012). As not much is known about the horse VEGFα gene, we retrieved horse VEGFα sequences from the Ensembl database and analyzed its molecular structure. We found that the gene had eight exons and was 696 bp long in horses (Figure 1A). When we examined the expression of VEGFα transcripts by RT-PCR in eight tissues, we found that all tissues ubiquitously expressed VEGFα transcripts. However, thyroid and lung tissue expressed the highest levels of VEGFα transcript whereas the skeletal muscle and appendix tissue expressed relatively low levels of VEGFα transcript (Figure 3).
Phylogenetic analysis of the VEGFα gene by neighbor-joining method revealed that equine VEGFα belongs to the same clade as the porcine VEGFα gene (Figure 2).
To better understand the evolution of the VEGFα gene, we analyzed synonymous (Ks) and non-synonymous (Ka) substitutions of VEGFα amino acids from ten species (Table 1). Pairwise comparison showed that Ks ranged from 0.168 to 2.112, whereas Ka ranged from 0.191 to 1.468. It is noted that Ks of equine VEGFα was the lowest with the pig gene (0.188) and the highest with the dog gene (2.112). The lowest Ks values were obtained between mouse and chimpanzee (0.092). The average of Ka and Ks were 0.839 and 0.689, respectively, resulting in a Ka/Ks ratio of greater than 1. This means that non-synonymous substitutions occurred more rapidly than synonymous substitutions. These results are based on the assumption that the protein-coding regions in the human genome have been under positive selection during evolution (Bustamante et al., 2005). These data indicated that positive selection occurred in the VEGFα genes during the evolutionary history of the horse.
To investigate the relationship between VEGFα gene expression level and time length after exercise, we performed qPCR analysis using blood leukocyte samples obtained before and after exercise (30 min, 60 min, 90 min, 120 min) from three horses (Figure 4A and B). Interestingly, the expression level of VEGFα was the highest after 60 min of exercise and then gradually decreased after upto 120 min. But the expression level of 120 min was slightly higher than that of 0 min (before exercise, p>0.05).
In horse, as there are few reliable biomarkers for recovery after exercise except creatine kinase (Linder et al., 2006) and lactate (Lindner et al., 2009), it is necessary to identify and evaluate markers for post exercise recovery. In this sense, RNA-seq based transcriptome analysis may be useful. In a previous study, horse VEGFα transcript increased significantly in response to exercise (Park et al., 2012) similar to humans suggesting a conserved mechanism for the regulation of VEGFα transcription. Therefore, we selected the equine VEGFα gene for further evaluation at the molecular level. A phylogenetic analysis showed that the horse VEGFα gene is grouped into the same clade as the pig gene and have undergone positive selection. In terms of expression, horse VEGFα was found to be ubiquitously expressed but regulated by exercise in blood leukocytes. Especially, expression kinetics obtained in this study showed that the transcript level gradual increased up to 60 min (p<0.05), and then remained at a slightly higher level than at time 0, which is not statistically significant (p>0.05). It is not certain how VEGFα expression is upregulated and reached the highest level after 60 min of exercise. One explanation would be that acute exercise may induce the hypoxia condition in leukocytes, which triggers hypoxia-inducible factor-1α (HIF-1α) activation. Activated HIF-1α will induce the transcriptional activation of VEGFα by binding to HIF-responsive elements which are located in the promoter region of VEGFα (Jiang et al., 1996). Therefore, it is worth examining equine HIF-1α expression in blood leukocytes after exercise to reveal the causal relationship between HIF-1α and VEGFα expression.
Nonetheless, the role of VEGFα in response to exercise and recovery is not clear in blood. It is likely that the circulating VEGFα facilitates mobilization of monocytes into injured areas after exercise, and promotes wound healing by stimulating endothelial cell proliferation and extracellular matrix changes as process of recovery (Rocha et al., 1988; Richardson et al., 1999). Additionally, vascular distribution and density increase in response to exercise in skeletal muscle as an adaptation, which accompanies increase VEGFα expression (Brodal et al., 1977). Further study is warranted to explore the relationship between expression levels of VEGFα in muscle and blood with physiological changes.
In summary, we analyzed molecular structure of equine VEGFα gene and performed phylogenetic analysis with homologues of other species, which revealed that VEGFα gene of horse belongs to the same clade with that of pig. Gene expression analysis using various horse tissues showed the ubiquitous expression of horse VEGFα gene. Also, it is of note that VEGFα expression in blood leukocytes is regulated in response to exercise, suggesting that horse VEGFα expression in leukocytes can be used as a biomarker for recovery after exercise.
ACKNOWLEDGMENTS
This work was supported by a grant from the Next- Generation BioGreen 21 Program (No. PJ008106, PJ008196), Rural Development Administration, Republic of Korea.
Figure 1
Gene structure and deduced amino acids sequences of equine VEGFα gene. (A) Structure of equine VEGFα gene. (B) Deduced amino acid sequences of equine VEGFα from mRNA sequences.
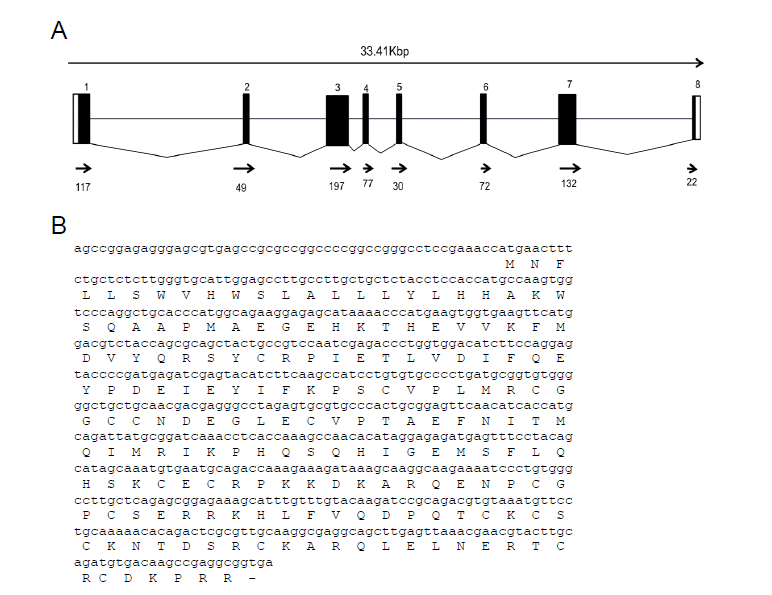
Figure 2
Phylogenetic tree of the VEGFα gene from ten different species using the neighbor-joining method.
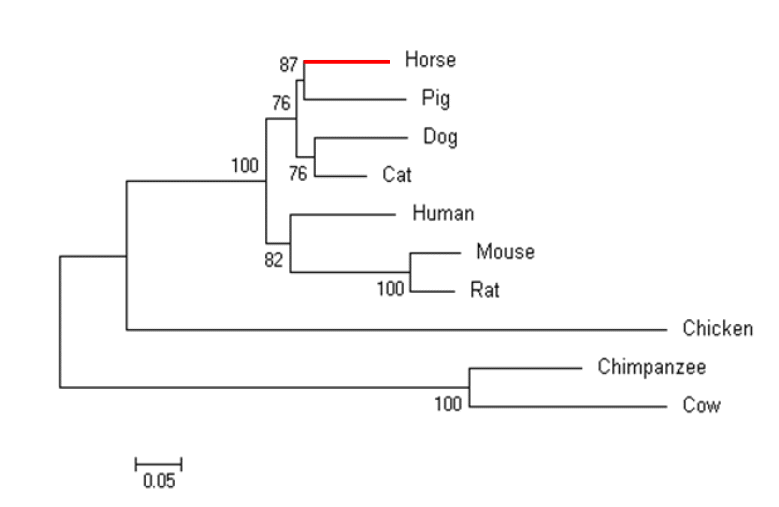
Figure 3
Expression levels of equine VEGFα gene by RT-PCR analysis in various horse tissues. RT-PCR products were visualized by gel electrophoresis. 1, skeletal muscle; 2, kidney; 3, thyroid; 4, lung; 5, appendix; 6, colon; 7, spinal cord; 8, heart. This is a representative result of three animals (n = 3). GAPDH gene was used as a control.
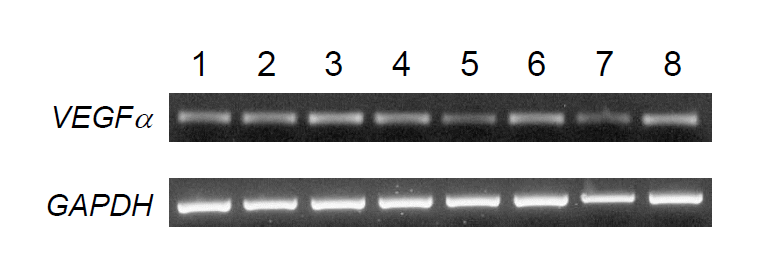
Figure 4
Differential expression of VEGFα gene in horse blood leukocytes after exercise. (A) RT-PCR of horse VEGFα gene in leukocytes of horse before exercise to post exercise at 120 min (a representative result of three horses). (B) Expression analysis of the VEGFα gene used to qPCR. Expression was calculated by 2−ΔΔCT method. 0 min, before exercise; 30 min, 30 min after exercise; 60 min, 60 min after exercise; 90 min, 90 min after exercise; 120 min, 120 min after exercise. This is a representative result of three animals (n = 3). GAPDH gene was used as a control. * p<0.05.
Table 1
Synonymous (Ks) and non-synonymous substitutions (Ka) per amino acid site in the VEGFα gene of ten species
REFERENCES
Auer DE, Ng JC, Hrdlicka J, Seawright AA. 1989. The elimination of injected superoxide dismutase from synovial fluid of the horse. Aust Vet J 66:117–119.


Bertone AL, Palmer JL, Jones J. 2001. Synovial fluid cytokines and eicosanoids as markers of joint disease in horses. Vet Surg 30:528–538.


Breen EC, Johnson EC, Wagner H, Tseng HM, Sung LA, Wagner PD. 1996. Angiogenic growth factor mRNA responses in muscle to a single bout of exercise. J Appl Physiol 81:355–361.


Brodal P, Ingjer F, Hermansen L. 1977. Capillary supply of skeletal muscle fibers in untrained and endurance-trained men. Am J Physiol Heart Circ Physiol 232:H705–H712.

Brutsaert TD, Gavin TP, Fu Z, Breen EC, Tang K, Mathieu-Costello O, Wagner PD. 2002. Regional differences in expression of VEGF mRNA in rat gastrocnemius following 1 hr exercise or electrical stimulation. BMC Physiol 2:8



Bustamante CD, Fledel-Alon A, Williamson S, Nielsen R, Hubisz MT, Glanowski S, Tanenbaum DM, White TJ, Sninsky JJ, Hernandez RD, Civello D, Adams MD, Cargill M, Clark AG. 2005. Natural selection on protein-coding genes in the human genome. Nature 437:1153–1157.


Essén-Gustavsson B, Lindholm A. 1985. Muscle fibre characteristics of active and inactive standard bred horses. Equine Vet J 17:434–438.


Firth EC. 2006. The response of bone, articular cartilage and tendon to exercise in the horse. J Anat 208:513–526.



Hinchcliff KW, Geor RJ. 2008. The Horse as an Athlete: A Physiological Overview. Equine exercise physiology: The science of exercise in the atheletic horse. Saunders/Elsevier; Edinburgh, UK, New York, USA: p. ixp. 463

Hyyppä S, Räsänen LA, Pösö AR. 1997. Resynthesis of glycogen in skeletal muscle from standardbred trotters after repeated bouts of exercise. Am J Vet Res 58:162–166.


Jiang BH, Rue E, Wang GL, Roe R, Semenza GL. 1996. Dimerization, DNA binding, and transactivation properties of hypoxia-inducible factor 1. J Biol Chem 271:17771–17778.


Lamprecht ED, Bagnell CA, Williams CA. 2008. Inflammatory responses to three modes of intense exercise in Standardbred mares-A pilot study. Comp Exer Physiol 5:115–125.

Lindner A, Signorini R, Brero L, Arn E, Mancini R, Enrique A. 2006. Effect of conditioning horses with short intervals at high speed on biochemical variables in blood. Equine Vet. J 38:Suppl 3688–92.

Lindner A, Mosen H, Kissenbeck S, Fuhrmann H, Sallmann HP. 2009. Effect of blood lactate-guided conditioning of horses with exercises of differing durations and intensities on heart rate and biochemical blood variables. J Anim Sci 87:3211–3217.


Livak K, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408.


Park KD, Park JS, Ko JS, Kim BC, Kim HS, Ann K, Do KT, Choi HS, Kim HM, Song SH, Lee SW, Kong HS, Yang YM, Jhun BH, Kim CH, Kim TH, Hwang SW, Bhak J, Lee JK, Cho BW. 2012. Whole transcriptome analyses of six thoroughbred horses before and after exercise using RNA-Seq. BMC Genomics 13:473



Petersen HH, Nielsen JP, Heegaard PM. 2004. Application of acute phase protein measurements in veterinary clinical chemistry. Vet Res 35:163–187.


Pösö AR, Essén-Gustavsson B, Persson SG. 1993. Metabolic response to standardbred trotters with red-cell hypervolemia. Equine Vet J 25:527–531.


Richardson RS, Wagner H, Mudaliar SR, Henry R, Noyszewski EA, Wagner PD. 1999. Human VEGF gene expression in skeletal muscle: Effect of acute normoxic and hypoxic exercise. Am J Physiol Heart Circ Physiol 277:H2247–2252.

Richardson RS, Wagner H, Mudaliar SRD, Saucedo E, Henry R, Wagner PD. 2000. Exercise adaptation attenuates VEGF gene expression in human skeletal muscle. Am J Physiol Heart Circ Physiol 279:H772–H778.


Roca J, Gavin TP, Jordan M, Siafakas N, Wagner H, Benoit H, Breen E, Wagner PD. 1998. Angiogenic growth factor mRNA responses to passive and contraction-induced hyperperfusion in skeletal muscle. J Appl Physiol 85:1142–1149.


Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425.






 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print





