 |
 |
| Anim Biosci > Volume 36(1); 2023 > Article |
|
Abstract
Objective
The objective of this study was to investigate the effects of feeding active dry yeast (ADY) and yeast culture (YC) on fecal bacterial community in finishing bulls fed high-concentrate diets in the same experimental environment.
Methods
Forty-five healthy finishing cattle (Simmental×Chinese Luxi yellow bulls; 24 months; 505±29 kg) were randomly divided into three groups: i) CON group (control group, only fed basal diet), ii) ADY group (fed basal diet + active dry yeast), and iii) YC group (fed basal diet + yeast culture). At the end of the trial, nine rectum fecal samples were randomly selected from each group for bacterial DNA sequencing.
Results
There was no difference among groups about alpha diversity indices (all p>0.05), including ACE, Chao 1, Shannon, and Simpson indices. Principal component analysis and non-metric multidimensional scaling analysis showed a high similarity among three groups. Compared with CON group, ADY and YC groups had greater relative abundance of c_Clostridia, o_Oscillospirales, and f_Oscillospiraceae, but lesser relative abundance of g_Megasphaera, and s_Megasphaera_elsdenii (all p<0.01). And, the relative abundances of p_Firmicutes (p = 0.03), s_Prevotella_sp (p = 0.03), o_Clostridiales (p<0.01), g_Clostridium (p<0.01), f_Caloramatoraceae (p<0.01), and f_Ruminococcaceae (p = 0.04) were increased in the ADY group. The PICRUSt2 prediction results showed that the metabolic pathways had no significant differences among groups (p>0.05). Besides, the relative abundance of c_Clostridia (r = 0.42), and f_Oscillospiraceae (r = 0.40) were positively correlated to average daily gain of finishing bulls (p<0.05).
Conclusion
Both of ADY and YC had no effect on diversity of fecal bacteria in finishing bulls, but the supplementation of ADY and YC can improve the large intestinal function in finishing bulls by increasing the abundance of cellulolytic bacteria and altering the abundance of lactic acid-utilizing bacteria.
The composition and function of cattle gastrointestinal microbiome plays a critical role in animal health and nutrient uptake [1]. Among them, bacteria play a dominant role in metabolomic activities of cattle [2]. To date, many studies have focused on rumen bacteria and their relationship with gastrointestinal microenvironment and production performance of cattle, but there are few studies focused on bacteria in the hindgut of beef cattle. The difference of bacterial communities between ruminant hindgut and rumen may be related to the structural and functional differences of the two digestive organs. Compared with the bacteria in rumen, the relative abundance of Bacteroidetes and Spirochaetes in the ruminant hindgut is lower, while Firmicutes and Proteobacteria is higher [3]. The hindgut microorganisms can degrade unutilized substances into volatile fatty acid to provide nutrition for the body [4], and some certain species of Bacteroidetes and Firmicutes may be related to the regulation of host behavior and intestinal immunity [5]. Therefore, the study of ruminant hindgut microorganisms is conducive to the overall understanding of ruminant gastrointestinal function.
Yeast preparations from Saccharomyces cerevisiae can be classified into two types according to the count of live yeast cells in products, which are active dry yeasts (ADY) and yeast culture (YC). Active dry yeasts which guarantee high number (>109 colony forming units/g) of live yeast cells are sold as 100% ADY, while YC are sold as entire culture medium with a small amount of live yeast cells. Although YC contains some residual viable yeast cells, which is not a substantial source of yeast biomass, and the effective components of YC are extracellular metabolites, such as peptides, alcohols, esters, and organic acids [6]. ADY and YC have been used in ruminants to favorably modify the ruminal environment and improve production performance. Nevertheless, published literature regarding ADY or YC did not show conclusive evidence that its supplementation is beneficial for animal performance at all times [7–9]. The action mechanism of ADY and YC on growth performance of ruminants has been explored for its scientific applications and product development [10,11]. So far, most studies have paid more attention on the effects of yeast preparations on rumen microorganisms [12,13], but little on hindgut microorganisms in finishing cattle [14]. At present, it has been demonstrated that ADY can reduce fecal Escherichia coli O157:H7 counts [15] and alter the dominant fecal bacteria at phylum and genus levels [16]. However, the effect of YC on hindgut bacteria community and different effects of ADY and YC on the fecal bacteria community in finishing bulls fed high-concentrate diets remain unclear. Therefore, the objectives of this study were to determine the alteration of fecal bacteria community of finishing bulls fed high-concentrate diets with ADY and YC supplementation using 16S rDNA sequencing. Growth performance, carcass traits, meat quality and blood indexes were reported previously [7,17].
The study received the approval of the Animal Ethics Committee of Yanbian University (Yanji, China) and carried out in accordance with animal welfare guidelines and regulations.
The experimental design was as described in our previous study [7]. Briefly, 45 healthy finishing cattle (Simmental× Chinese Luxi yellow bulls; 24 months; with initial body weight of 505±29 kg) were randomly divided into three groups: i) CON group (control group, only fed a basal diet), ii) ADY group (fed basal diet + active dry yeasts preparation (Levucell SC, S. cerevisiae CNCM1-1077; white; >0.8×1010 CFU/g; 0.8 g/head/d), and iii) YC group (fed basal diet + yeast culture preparation (Diamond V XP, Cedar Rapids, IA, USA; 50 g/head/d). The basal diet was a total mixed ration with concentrate to forage ratio of 70:30 (as shown in Supplementary Table S1). The finishing bulls were fed twice a day at 05:00 and 17:00 for 98 days. At the end of the trial and before the morning feeding, nine cattle were randomly selected from each group for rectal fecal sampling by using sterile surgical gloves. One gram of feces was subsampled from each mixed fecal sample, snap frozen in liquid nitrogen and stored at −80°C for DNA extraction.
Total bacterial DNA was extracted from 125 mg fecal sample using a TGuide S96 Magnetic Soil/Stool DNA Kit (Tiangen Biotech Co., Ltd, Beijing, China). DNA sequencing was performed as described previously [18]. Briefly, DNA purity and concentration were determined with a Synergy HTX multi-mode reader (Gene Company Limited, Hong Kong, China) and DNA integrity was assessed by 1.8% agarose gel electrophoresis. First-round tailed polymerase chain reaction (PCR) amplification was performed as detailed in previous research [19]. The full-length 16S rDNA sequence was amplified using the universal primers: 27F (5′-AGRGTTTGAT YNTGGCTCAG-3′) and 1492R (5′-TASGGHTACCTTGT TASGACTT-3′). The second-round tailed PCR reaction system contained the barcode primer pair (3 μL), genomic DNA (1.5 μL), nuclease-free water (10.5 μL), and KOD OneTM PCR Master Mix (15 μL). The cycling parameters were as follows: initial denaturation for 2 min at 95°C, then 98°C for 10 s, 55°C for 30 s, and 72°C for 90 s, for 25 cycles, and a final extension for 5 min at 72°C. PCR amplification products were detected by Qubit4 fluorometer (Thermo Fisher, New York, USA) and 1.8% agarose gel electrophoresis, before purification, quantification, and homogenization to create a sequence library. The marker genes were sequenced by single molecule real-time sequencing using a HiSeq 2500 system PacBio Sequel II system (Pacific Biosciences, Menlo Park, CA, USA).
Analysis of sequence data followed a protocol described previously [19]. Effective reads were obtained by filtering the raw reads using Trimmomatic (v.0.33), identification and removal of primer sequences by cutadapt (v.1.9.1), splicing of high-quality reads by FLASH (v.1.2.7), and the identification and removal of potential chimera using the UCHIME algorithm. Operational taxonomic units (OTUs) were obtained by clustering reads at 97.0% similarity level using Usearch (v.10.0). Taxonomic annotation of OTUs based on the SILVA database (Release 132) used the naive Bayes classifier. Species abundance at phylum and genus levels was generated by QIIME2 (v.2020.6) and mapped by R (v.3.3.2). The alpha diversity indices (Chao1, ACE, Shannon, and Simpson) were evaluated using QIIME2 (v.2020.6). The alpha diversity data were analyzed using one-way analysis of variance with Dunnett T3 test. Statistical significance was set at p<0.05. Shannon curves and species accumulation curves (OTU level) were created using Mothur software and R (v.3.3.2). QIIME2 (v.2020.6) was used to determine beta diversity, the Bray Curtis algorithm to calculate the distance among samples to obtain the beta value, and three-dimensional principal component analysis (3D PCA) and non-metric multidimensional scaling analysis (NMDS) for analysis of beta diversity. Species for statistical differences among groups were analyzed using linear discriminant analysis (LDA) with LDA scores >4. Functional gene prediction analysis was carried out according to previous research [20]. Briefly, characteristic sequences were annotated using PICRUSt2 and matched with the Kyoto encyclopedia of genes and genomes database (KEGG) to predict the functional gene composition of a sample. STAMP software was used to carry out t-tests on functional abundances among groups.
We previously reported that the ADY supplementation can significantly improve the final weight (FW), dietary dry matter intake (DMI) and average daily gain (ADG), and both supplementation of ADY and YC can significantly improve the level of serum ghrelin and serum triglyceride (TG) of finishing bulls (Supplementary Table S2, 3) [7,17]. The correlations between differential fecal bacteria and FW, DMI, ADG, serum ghrelin and serum TG were calculated by Spearman’s rank correlation analysis with p value <0.05 being considered as significant.
In the present study, a total of 149,858 circular consensus sequences (CCS) were obtained by identifying barcodes and average sequence efficiency (effective CCS/Raw CCS) exceeded 93% in the sequencing results from twenty-seven fecal samples in finishing bulls by sequencing the 16S rDNA genes (Supplementary Figure S1). The results of the OTUs were used to create Venn diagrams, showing the numbers of microbes and variances of different groups of finishing bulls (Figure 1). The numbers of total OTUs in CON, ADY and YC groups were 381, 328, and 364, respectively. The number of mutual OTUs from the three groups was 278, representing 63.18% of all OTUs.
Beta diversity was used to compare the microbial community in different samples by calculating the 3D-PCA and NMDS. As shown in Figure 2, the 3D-PCA and NMDS plots based on the Bray-Curtis distance matrix showed that the points representing fecal microorganisms in the three groups were not independent, which indicated that the community structure of large intestinal bacteria had no difference among treatments.
Species relative abundance accumulation curve and the Shannon index rarefaction curves indicated that all samples provided sufficient OTU coverage to accurately describe the bacterial composition of each treatment (Figure 3A and 3B). In order to measure the diversity within the microbial community of each sample, we compared alpha diversity of the fecal bacteria in all samples by calculating the ACE, Chao1, Shannon, and Simpson indices (Figure 3C, 3D, 3E, and 3F). We found that the Simpson index in ADY group had an increased tendency compared with CON group (p = 0.07), but no significant differences were observed (Figure 3F).
At the phylum level, Firmicutes (CON vs ADY vs YC = 32.19% vs 43.18% vs 34.13%), Proteobacteria (CON vs ADY vs YC = 33.78% vs 15.01% vs 23.88%), and Bacteroidota (CON vs ADY vs YC = 15.68% vs 24.41% vs 25.76%) predominated in the hindgut of all groups, and occupied more than 80% of the microbial community (Figure 4A). The dominant bacterial genera identified in all finishing bulls’ fecal bacteria were Succinivibrio, Prevotella, Treponema, Bact, Roseburia, and Anaerovibrio (Figure 4B). Figure 4 also shows that the supplement of ADY or YC had altered abundances of specific bacterial taxa at the phylum and genus level. Hence, different types of yeast production could dissimilarly promote the growth of certain bacteria in the rectum of finishing bulls.
Furthermore, the significantly different abundant fecal bacteria between the 2 groups were identified through LEfSe analysis and LDA. Interestingly, compared with CON group, ADY and YC groups had greater relative abundance of c_Clostridia, o_Oscillospirales, and f_Oscillospiraceae, but lesser relative abundance of g_Megasphaera, and s_Megasphaera_elsdenii (all p<0.01) (Figure 5A and 5B). Besides, the relative abundance of f_Veillonellaceae (p = 0.01) and s_Lactobacillus_porci (p = 0.03) were decreased in ADY and YC groups (Figure 5A and 5B), respectively. And, the relative abundances of p_Firmicutes (p = 0.03), s_Prevotella_sp (p = 0.03), o_Clostridiales (p<0.01), f_Caloramatoraceae (p<0.01), g_Clostridium (p<0.01), and f_Ruminococcaceae (p = 0.04) were increased in the ADY group (Figure 5A). Compared with the ADY group, the relative abundances of g_Treponema (p<0.01), o_Spirochaetales (p<0.01), p_Spirochaetota (p<0.01), c_Spirochaetia (p<0.01), f_Spirochaetaceae (p<0.01), c_Clostridia (p<0.01), s_Candidatus_Treponema_suis (p = 0.01), and o_Clostridiales (p = 0.02) in YC group were decreased (Figure 5C).
The PICRUSt2 software was used to predict the metabolic functions of the identified bacterial 16S rDNA genes by using KEGG database. The metabolic pathways had no significant differences (p>0.05) between YC and CON groups (Figure 6B). Interestingly, the relative abundance of membrane transport and signal transduction genes were more abundant in ADY group than that in CON group (Figure 6A) or YC group (Figure 6C) (p<0.05). Besides, the functional genes of fecal bacteria in all groups were mainly associated with the following metabolic pathways: global and overviews maps, carbohydrate metabolism, amino acid metabolism, metabolism of cofactors and vitamins, nucleotide metabolism, translation, energy metabolism (in order from high to low abundance) (Supplementary Figure S2).
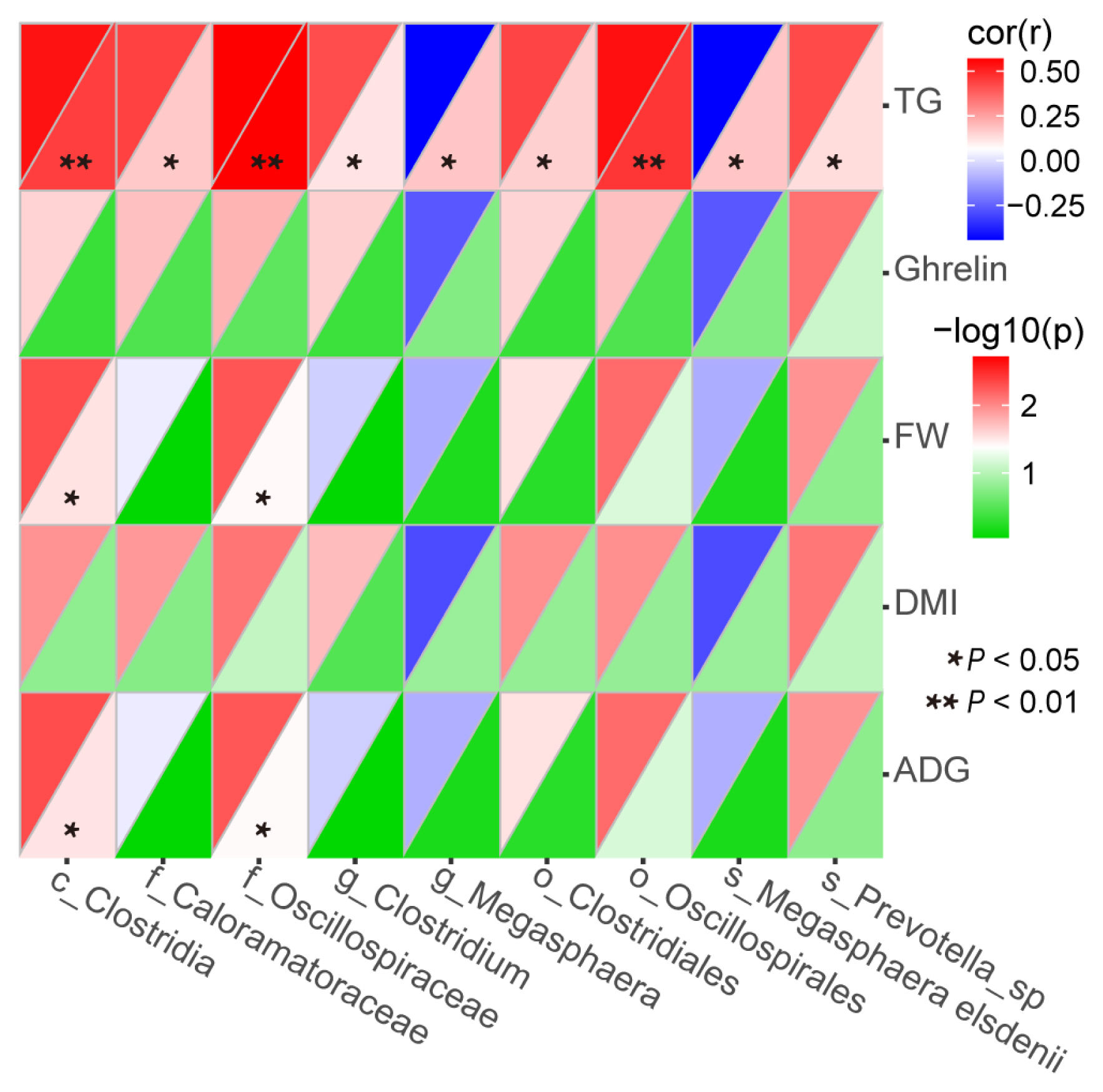
As shown in Figure 7, the relative abundance of c_Clostridia (r = 0.53; p<0.01), f_Caloramatoraceae (r = 0.44; p<0.05), f_Oscillospiraceae (r = 0.57; p<0.01), g_Clostridium (r = 0.42; p<0.05), o_Clostridiales (r = 0.43; p<0.05), o_Oscillospirales (r = 0.54; p<0.01) and s_Prevotella_sp (r = 0.42; p<0.05) were positively correlated with serum TG, whereas the relative abundance of g_Megasphaera (r = −0.44; p<0.05) and s_Megasphaera_elsdenii (r = −0.44; p<0.05) were negatively correlated. Meanwhile, the relative abundance of c_Clostridia (r = 0.42; r = 0.42), and f_Oscillospiraceae (r = 0.40; r = 0.40) were positively correlated with FW and ADG (all p<0.05).
In the present study, we evaluated the impact of different types of yeast preparations (ADY and YC) on the fecal bacteria community of finishing bulls based on the high-throughput sequencing technology. Some studies reported that ADY alters the beta-diversity of rumen samples of beef steers [21]. Through the analysis of the diversity indices of samples in three groups, we found that the supplement of ADY and YC did not alter the fecal bacteria diversity in finishing bulls fed high-concentrate diets. This is consisted with Ran et al [16], who reported that neither ruminal protected nor non-protected ADY can change the structure of fecal bacteria of beef cattle. As known to us, yeast preparations can provide peptides, amino acids, vitamin and trace minerals to stimulate the growth of rumen microorganism [22]. Besides, live yeast cells of ADY can also scavenge oxygen to create an anaerobic ruminal environment for the growth and multiplication of anaerobic bacteria, which is one of the important reasons why ADY affect the structure of rumen bacterial community [23]. But a factor of scavenge oxygen by ADY supplementation may not be the key for bacteria in the ruminant anaerobic hindgut. In addition, it is also possible that almost all the nutrients of growth factors in yeast preparations are utilized in rumen microorganisms, and the contents of various nutritional factors in chyme are not different enough to cause changes in the structure of hindgut flora.
Even so, the LEfSe analysis results indicated that yeast metabolites in yeast preparations effectively regulate the relative abundance of certain fecal bacteria in the current study. For example, the supplementation of ADY and YC significantly increased the relative abundance of c_Clostridia, o_Oscillospirales, and f_Oscillospiraceae. As a strict anaerobe, Clostridia has the ability to ferment complex plant carbohydrates [24]. Clostridia species produce short chain fatty acids (e.g. butyrate), mucin and antimicrobial peptides to provide essential nutrients and energy and enhance epithelial barrier integrity [25]. Besides, the increase of butyrate-producing bacteria - Clostridia suppresses the growth of aerobic Salmonella by increasing butyrate concentration and decreasing epithelial oxygenation in mammalian intestine [26]. Of course, there are some potentially toxic and pathogenic strains in Clostridium genus [27], but beneficial strains dominate after supplementing yeast preparations in the current study, judging from the physiological state, growth performance and plasma indexes of cattle (Supplementary Table S2, S3) [7,17]. As a common gut microbiota, Oscillospira is a rarely cultivated bacterial genus, which can utilize glucuronate and produce all kinds of short-chain fatty acids, especially butyrate [28]. Generally, Oscillospira is also positively associated with fiber content of diets [29]. That means the supplementing of yeast metabolites can enhance the intestinal digestion of fiber polysaccharide in finishing bulls. It is also reported that the increase abundance of Oscillospira may aggravate constipation in human [30]. Hence, the addition of yeast preparation can effectively reduce the duration of calf diarrhea [31,32], which may be also related to the increased abundance of this organism. Oscillospira consumes glucose, ethanol, and lactic acid in the culture medium for growth and multiplication, which may be the reason why the bacteria in hindgut supplemented with yeast preparation is more conducive to adapt to high grain diet [33]. This is also confirmed by our study, in which a positive correlation was observed between c_Clostridia, f_Oscillospiraceae and serum TG, FW, ADG in finishing bulls, indicating that the relative abundance of this bacteria in the rectum increased according to the growth performance. However, the reduced relative abundance of g_Megasphaera and s_Megasphaera_elsdenii were also observed after 98 days of supplementation with ADY and YC. Megasphaera elsdenii belongs to family Veillonellaceae and is the common lactate-utilising bacteria in the rumen of grain-fed cattle [34]. the supplementation of ADY also decreased the relative abundance of f-Veillonellaceae. Ogunade et al [35] demonstrated that yeast preparations have the ability to increase the abundance of carbohydrate digesting bacteria and lactate-utilising bacteria. Thus, we speculate that the increased relative abundance of Oscillospira may compensate for the decrease of Megasphaera in the hindgut of finishing bulls fed high-concentrate diets. The mechanism leading to this strange phenomenon still needs further studies. Besides, the relative abundance of s_Lactobacillus_porci was decreased after supplementing YC. This agrees with Lesmeister et al [36], who demonstrated YC could restrain the activities of lactate-producing bacteria. With glucose as a carbon source, Lactobacillius can produce lactic acid [37]. Hence, reducing the production of lactic acid by decreasing the relative abundance of s_Lactobacillus_porci could prevent hindgut acidosis [38,39]. Our observations suggest that both ADY and YC can regulate the relative abundance of cellulolytic bacteria and lactic acid-utilizing bacteria in the hindgut and potentially improve the adaptability of the intestine to high-energy diet in finishing bulls.
Besides, the supplementation of ADY also increased the relative abundances of p_Firmicutes, s_Prevotella_sp and f_Ruminococcaceae. These results are consistent with Ran et al [16], who found that both ruminal protected and non-protected ADY can effectively improve the relative abundance of phylum_Firmicutes and genus_Prevotella. Irrespective of diet fed to animals, the phylum Firmicutes would probably be the most dominant in high grain diet [40]. Furthermore, increased relative abundance of Firmicutes enhanced energy harvesting in bovine and played a crucial role in increasing fat deposition in cows and feed efficiency in steers [41,42]. The polysaccharide-degrading Prevotellaceae bacterium has greater relative abundance in the rumen of cows fed high-concentrate diets, which can utilize and convert lactic acid into propionic acid [43]. As one of the most abundant families from the order Clostridiales in gut, the Ruminococcaceae has abundant genes encoding key carbohydrate-active enzymes, and could degrade complex plant material, including cellulose and hemicellulose, to produce short chain fatty acids (mainly acetate, butyrate, and propionate) [44]. These results suggest that ADY supplementation potentially improves the metabolism of polysaccharide in the large intestine of finishing bulls fed high-concentrate diets.
Furthermore, compared with supplementing ADY, the decreased relative abundances of o_Clostridiales, c_Clostridia, p_Spirochaetota, c_Spirochaetia, o_Spirochaetales, g_Treponema, f_Spirochaetaceae, and s_Candidatus_Treponema_suis were observed in the hindgut with supplementation of YC. As mentioned above, the great majority the Clostridia have a beneficial and commensal relationship with the host, although some of them are pathogenic. Spirochaetaceae (mainly the genus Treponema spp.) has the ability of fiber degradation [45,46]. Although Treponema include harmless commensal bacterial species, they are primarily known as potential pathogens [47]. Candidatus Treponema suis is known to cause colitis by invading the surface epithelium and the mucosa in pigs [48]. One species of genus Treponema (T. brennaborense), whose sequence is 90% similar to Candidatus Treponema suis, was found to be associated with digital dermatitis in dairy cows [49]. However, we did not find diarrhea, inflammation or other health problems in finishing bulls during the trial [7,17]. Moreover, most of researches reported that both ADY and YC also could reduce the diarrhea rate and improve immunocompetence and fecal scores of dairy calves [31,50]. Thus, the fiber degradation ability of the gut microbiota supplemented YC seems weaker than that of ADY supplementation in the current experimental condition, but it is difficult to distinguish which is more beneficial to the large intestinal health.
The fecal microbiota of beef cattle plays an important role in physiology, nutrition, health, and productivity. In the current study, phyla Firmicutes, Proteobacteria, and Bacteroidota predominated and occupied more than 80% of the fecal bacterial community in all finishing bulls, which is consistent with previous reports [16,51–53]. Succinivibrio was the first dominant bacterial group at genus level in this study. However, Ran et al [16] found Prevotella was the first dominant genus in fecal bacteria in cattle fed with high-grain diets. This may have been caused by different breed and dietary components. Furthermore, Hernandez-Sanabria et al [54] revealed that Succinivibrio were abundant in rumen of dairy cattle fed with high-concentrate and were positively correlated with feed efficiency and productivity. This indicates that the most dominant bacteria at genus level are highly variable in hindgut of finishing bulls. Although the supplementation of ADY and YC altered the relative abundance of certain fecal flora, the PICRUSt2 prediction results showed that it did not change the metabolic pathways of large intestinal bacteria, including carbohydrate metabolism, amino acid metabolism, metabolism of cofactors and vitamins, nucleotide metabolism, translation, energy metabolism. This indicates that different fecal bacteria can have similar functions in the hindgut ecosystem, and the gastrointestinal bacterial flora may achieve its stability through functional redundancy [55]. Interestingly, the relative abundance of membrane transport and signal transduction genes were more abundant in hindgut bacterial community added with ADY, which showed that feeding finishing bulls with ADY had potential to maintain the stability of intestinal flora by enhancing the response ability of intestinal bacteria to external stimuli [56].
In conclusion, both of ADY and YC had no effect on diversity of fecal bacteria in finishing bulls, but the supplementation of ADY and YC can alter the relative abundance of some cellulolytic bacteria and lactic acid-utilizing bacteria in the hindgut, and YC had a weaker effect than ADY in the current experimental condition. Furthermore, the relative abundance of class_Clostridia and family_Oscillospiraceae were positively correlated with average daily gain of finishing bulls receiving ADY and YC. The findings contribute to a better understanding of potential effects of yeast preparations on finishing bulls fed high-concentrate diets.
Notes
SUPPLEMENTARY MATERIAL
Supplementary file is available from: https://doi.org/10.5713/ab.22.0215
Supplementary Table S1. Ingredient and nutritional composition of basal diets (% of dry matter)
ab-22-0215-suppl1.pdf
Supplementary Table S2. Effect of dietary supplementation of active dry yeast and yeast culture on growth performance and blood indexes of finishing bulls (n = 15)
ab-22-0215-suppl2.pdf
Supplementary Table S3. Effect of dietary supplementation active dry yeasts (ADY) and yeast culture (YC) on blood immunoglobulin and blood hormone in finishing bulls (n = 15)
ab-22-0215-suppl3.pdf
Figure 1
Venn diagram of number of operational taxonomic units of rectum fecal bacteria in finishing bulls. CON, control group (n = 9); ADY, active dry yeast group (n = 9); YC, yeast culture group (n = 9).
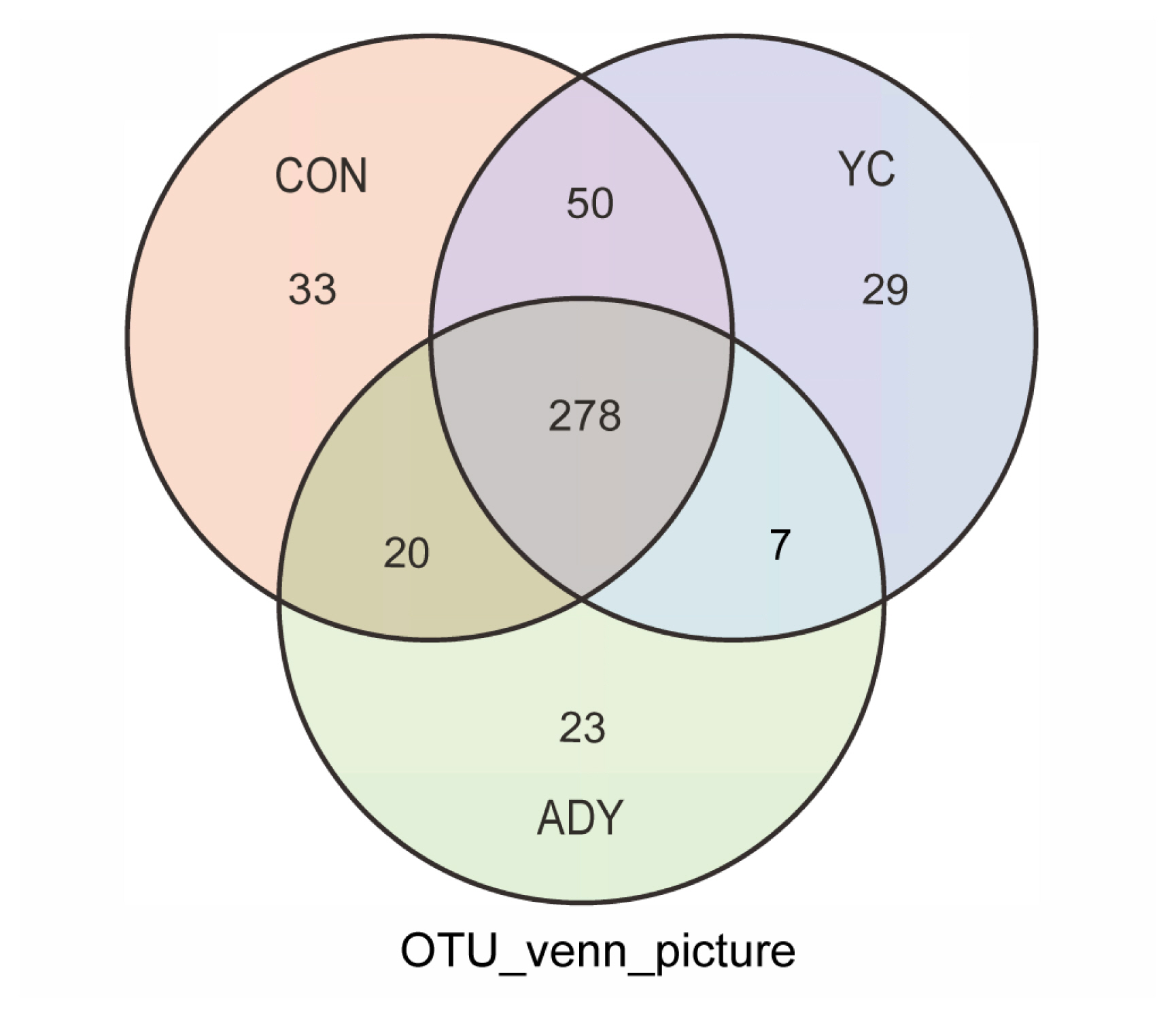
Figure 2
Beta diversity analysis of rectum fecal bacteria through (A) three-dimensional principal component analysis (3D-PCA) and (B) non-metric multidimensional scaling analysis (NMDS). CON, control group (n = 9); ADY, active dry yeast group (n = 9); YC, yeast culture group (n = 9).
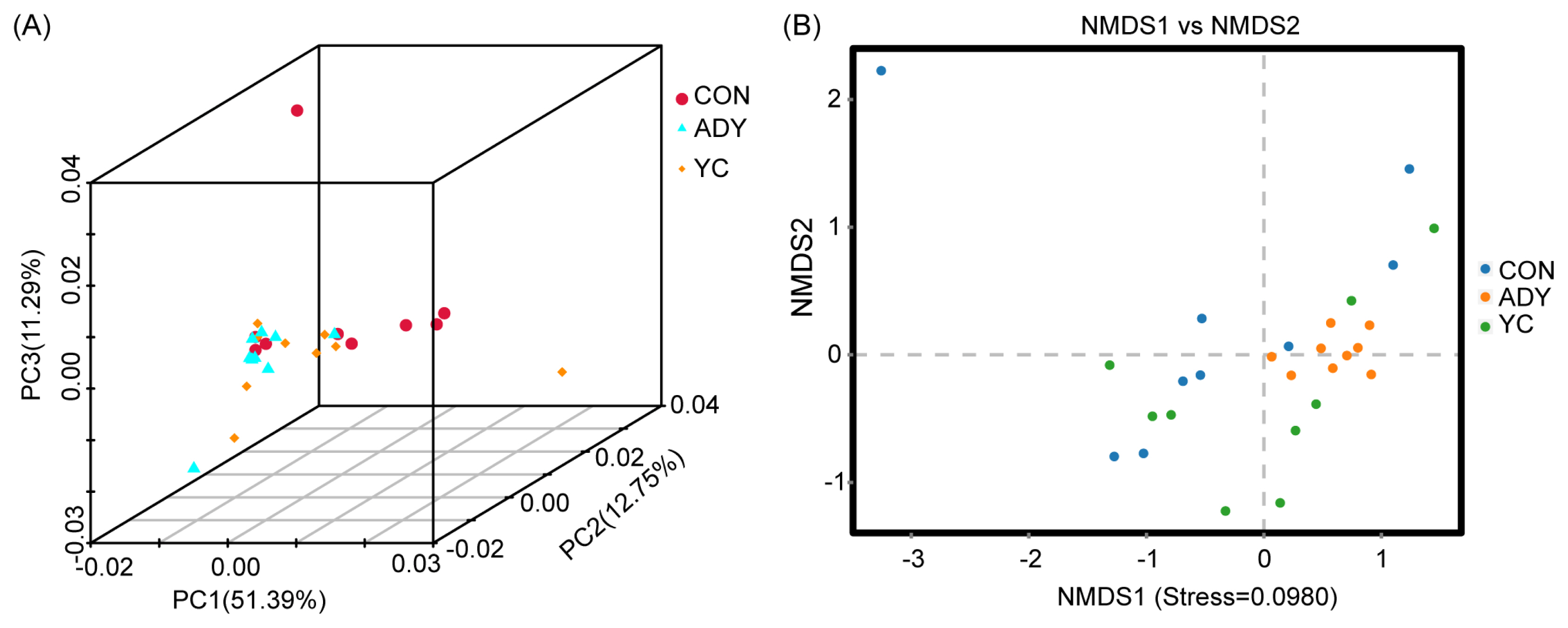
Figure 3
Alpha diversity of the rectum fecal bacteria in finishing bulls. (A) Species relative abundance accumulation curve, (B) the Shannon index rarefaction curves, (C) Abundance-based coverage estimator (ACE) index, (D) Chao 1 index, (E) Shannon index and (F) Simpson index. A single red box reflects the total number of species contained in the sample, and the total red box constitutes a cumulative curve. A single green box reflects the number of common species in the sample; The total green box constitutes the common quantity curve. CON, control group (n = 9); ADY, active dry yeast group (n = 9); YC, yeast culture group (n = 9).
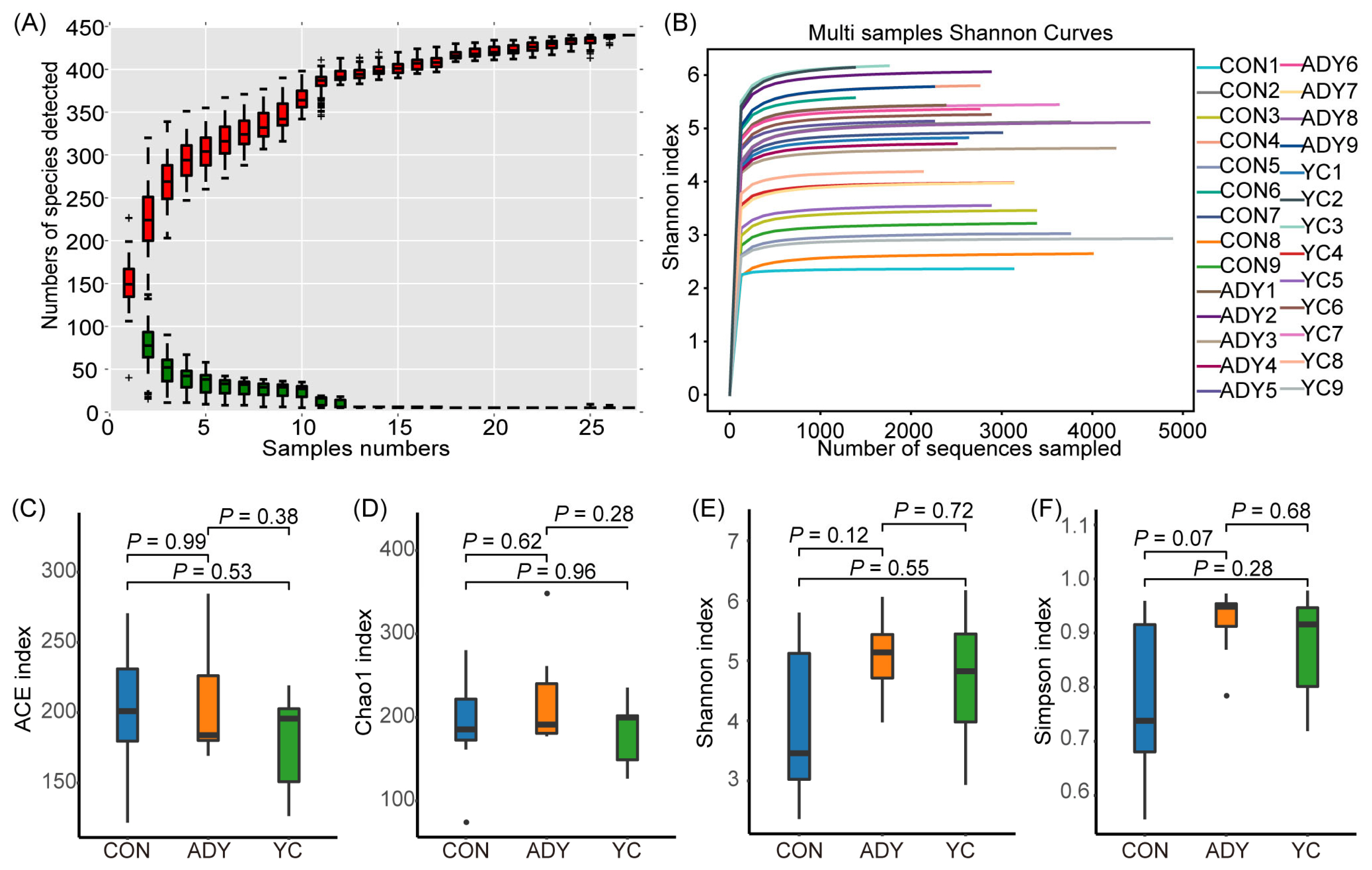
Figure 4
The rectum fecal bacterial community compositions at (A) phylum (all) and (B) genus (top 15) level. Taxonomy was assigned using the SILVA database version 132. The different colors of the bars represent different species, and the length of the bars represents the proportion of the species. CON, control group (n = 9); ADY, active dry yeast group (n = 9); YC, yeast culture group (n = 9).
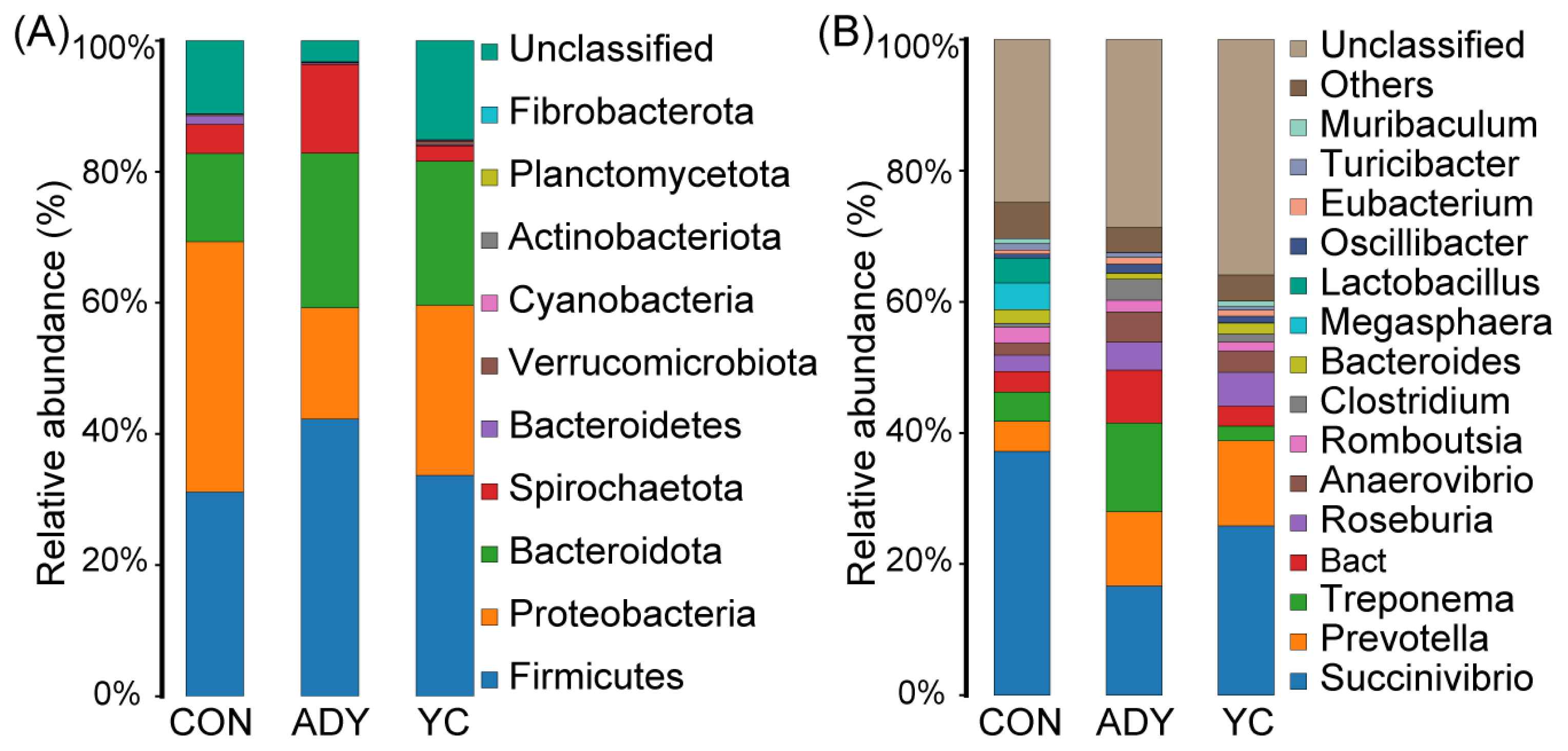
Figure 5
The linear discriminant analysis effect size (LEfSe) analysis of differential fecal bacteria. Linear discriminant analysis (LDA) bar showed the impact of the abundance of each species on the difference in (A) ADY vs CON, (B) YC vs CON, and (C) ADY vs YC. p-Value <0.05 and LDA score >4 were defined as significant difference. p, phylum; c, class; o, order; f, family; g, genus; s, species. CON, control group (n = 9); ADY, active dry yeast group (n = 9); YC, yeast culture group (n = 9).
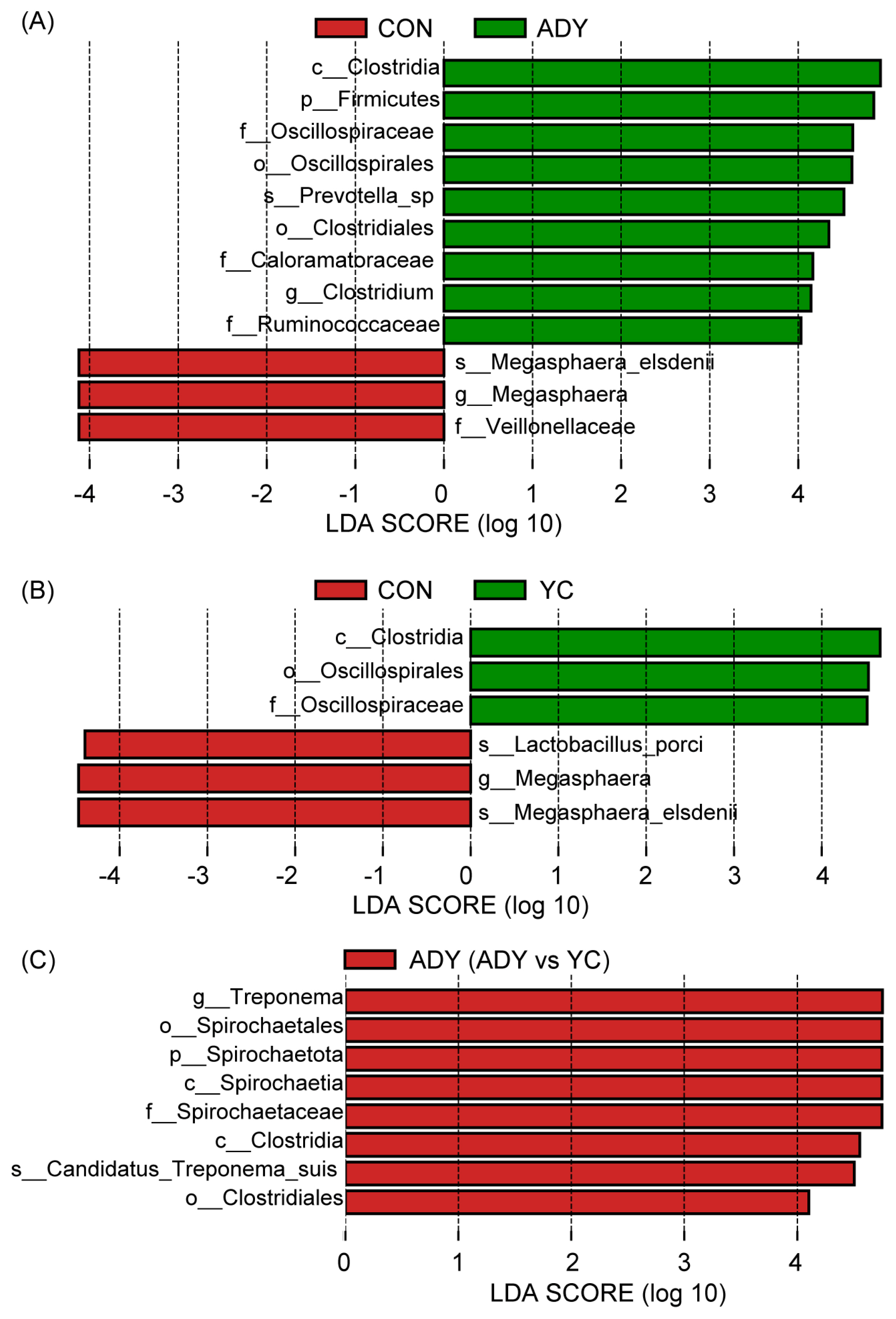
Figure 6
Comparison of predicted Kyoto encyclopedia of genes and genomes database (KEGG) functions of fecal bacteria at level 2 by using PICRUSt2 (top10 and p-value (corrected) <1). Functional classification of (A) ADY vs CON, (B) YC vs CON, and (C) ADY vs YC. CON, control group (n = 9); ADY, active dry yeast group (n = 9); YC, yeast culture group (n = 9).
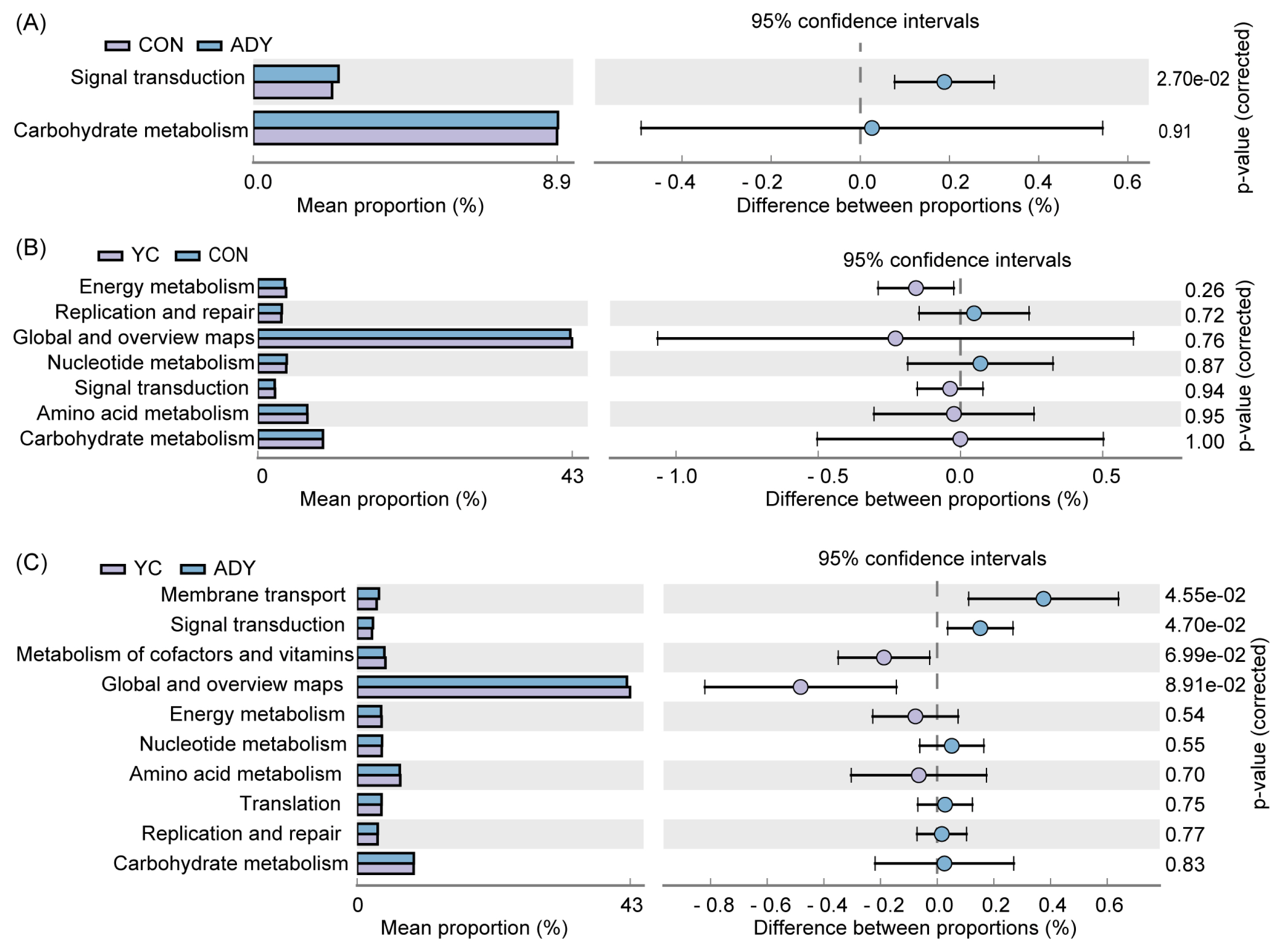
Figure 7
Spearman’s rank correlations between differential fecal bacteria and final weight, dietary dry matter intake, average daily gain, serum ghrelin and serum triglyceride. FW, final weight; DMI, dietary dry matter intake; ADG average daily gain; and TG, serum triglyceride. Spearman’s rank correlation coefficient (r) was from −1 to 1. r>0 and <0 represented a positive and negative correlation, respectively. The (r) value denoted the degree of correlation between variables. Only the bacteria with a relative abundance of 1%, or higher, in at least one sample were considered.
REFERENCES
1. Malmuthuge N, Guan LL. Understanding host-microbial interactions in rumen: searching the best opportunity for microbiota manipulation. J Anim Sci Biotechnol 2017; 8:8
https://doi.org/10.1186/s40104-016-0135-3



2. Van Soest PJ. Nutritional ecology of the ruminant. Ithaca, NY, USA: Cornell University Press; 1994.
3. Godoy-Vitorino F, Goldfarb KC, Karaoz U, et al. Comparative analyses of foregut and hindgut bacterial communities in hoatzins and cows. ISME J 2012; 6:531–41.
https://doi.org/10.1038/ismej.2011.131



4. Mao S, Zhang R, Wang D, Zhu W. The diversity of the fecal bacterial community and its relationship with the concentration of volatile fatty acids in the feces during subacute rumen acidosis in dairy cows. BMC Vet Res 2012; 8:237
https://doi.org/10.1186/1746-6148-8-237



5. Ashida H, Ogawa M, Kim M, Mimuro H, Sasakawa C. Bacteria and host interactions in the gut epithelial barrier. Nat Chem Biol 2011; 8:36–45.
https://doi.org/10.1038/nchembio.741


6. Shurson GC. Yeast and yeast derivatives in feed additives and ingredients: Sources, characteristics, animal responses, and quantification methods. Anim Feed Sci Technol 2018; 235:60–76.
https://doi.org/10.1016/j.anifeedsci.2017.11.010

7. Geng CY, Ren LP, Zhou ZM, Chang Y, Meng QX. Comparison of active dry yeast (Saccharomyces cerevisiae) and yeast culture for growth performance, carcass traits, meat quality and blood indexes in finishing bulls. Anim Sci J 2016; 87:982–8.
https://doi.org/10.1111/asj.12522


8. Swyers KL, Wagner JJ, Dorton KL, Archibeque SL. Evaluation of Saccharomyces cerevisiae fermentation product as an alternative to monensin on growth performance, cost of gain, and carcass characteristics of heavy-weight yearling beef steers. J Anim Sci 2014; 92:2538–45.
https://doi.org/10.2527/jas.2013-7559


9. Hinman DD, Sorensen SJ, Momont PA, Albin R, Cole NA. Effect of yeast culture on steer performance, apparent diet digestibility, and carcass measurements when used in a barley and potato finishing diet. Prof Anim Sci 1998; 14:173–7.
https://doi.org/10.15232/S1080-7446(15)31819-2

10. Geng CY, Feng X, Luan JM, Ji S, Jin YH, Zhang M. Improved tenderness of beef from bulls supplemented with active dry yeast is related to matrix metalloproteinases and reduced oxidative stress. Animal 2022; 16:100517
https://doi.org/10.1016/j.animal.2022.100517


11. Wiedmeier RD, Arambel MJ, Walters JL. Effect of yeast culture and Aspergillus oryzae fermentation extract on ruminal characteristics and nutrient digestibility. J Dairy Sci 1987; 70:2063–8.
https://doi.org/10.3168/jds.S0022-0302(87)80254-0


12. Shen Y, Wang H, Ran T, Yoon I, Saleem AM, Yang W. Influence of yeast culture and feed antibiotics on ruminal fermentation and site and extent of digestion in beef heifers fed high grain rations. J Anim Sci 2018; 96:3916–27.
https://doi.org/10.1093/jas/sky249



13. Amin AB, Mao S. Influence of yeast on rumen fermentation, growth performance and quality of products in ruminants: a review. Anim Nutr 2021; 7:31–41.
https://doi.org/10.1016/j.aninu.2020.10.005



14. Bach A, López-García A, González-Recio O, et al. Changes in the rumen and colon microbiota and effects of live yeast dietary supplementation during the transition from the dry period to lactation of dairy cows. J Dairy Sci 2019; 102:6180–98.
https://doi.org/10.3168/jds.2018-16105


15. Ran T, Shen Y, Saleem AM, AlZahal O, Beauchemin KA, Yang W. Using ruminally protected and nonprotected active dried yeast as alternatives to antibiotics in finishing beef steers: growth performance, carcass traits, blood metabolites, and fecal Escherichia coli. J Anim Sci 2018; 96:4385–97.
https://doi.org/10.1093/jas/sky272



16. Ran T, Jiao P, AlZahal O, et al. Fecal bacterial community of finishing beef steers fed ruminally protected and non-protected active dried yeast. J Anim Sci. 2020. 98:skaa058
https://doi.org/10.1093/jas/skaa058



17. Geng CY, Ji S, Jin YH, et al. Comparison of blood immunity, antioxidant capacity and hormone indexes in finishing bulls fed active dry yeast (Saccharomyces cerevisiae) and yeast culture. Int J Agric Biol 2018; 20:2561–8.
https://doi.org/10.17957/IJAB/15.0822

18. Klemetsen T, Willassen NP, Karlsen CR. Full-length 16S rRNA gene classification of Atlantic salmon bacteria and effects of using different 16S variable regions on community structure analysis. Microbiology Open 2019; 8:e898
https://doi.org/10.1002/mbo3.898



19. Yuan X, Zhang X, Liu X, et al. Comparison of gut bacterial communities of grapholita molesta (Lepidoptera: Tortricidae) reared on different host plants. Int J Mol Sci 2021; 22:6843
https://doi.org/10.3390/ijms22136843



20. Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 2014; 30:3123–4.
https://doi.org/10.1093/bioinformatics/btu494



21. Ogunade I, Schweickart H, McCoun M, Cannon K, McManus C. Integrating 16S rRNA sequencing and LC-MS-based metabolomics to evaluate the effects of live yeast on rumen function in beef cattle. Animals 2019; 9:28
https://doi.org/10.3390/ani9010028



22. Büchl NR, Hutzler M, Mietke-Hofmann H, Wenning M, Scherer S. Differentiation of probiotic and environmental Saccharomyces cerevisiae strains in animal feed. J Appl Microbiol 2010; 109:783–91.
https://doi.org/10.1111/j.1365-2672.2010.04705.x


23. Vohra A, Syal P, Madan A. Probiotic yeasts in livestock sector. Anim Feed Sci Technol 2016; 219:31–47.
https://doi.org/10.1016/j.anifeedsci.2016.05.019

24. Jones DT, Woods DR. Acetone-butanol fermentation revisited. Microbiol Rev 1986; 50:484–524.
https://doi.org/10.1128/mr.50.4.484-524.1986



25. Lewis JD, Ruemmele FM, Wu GD. Nutrition, gut microbiota and immunity: therapeutic targets for IBD. Concluding remarks. Nestle Nutr Inst Workshop Ser 2014; 79:161–2.
https://doi.org/10.1159/000360721


26. Rivera-Chávez F, Zhang LF, Faber F, et al. Depletion of butyrate-producing clostridia from the gut microbiota drives an aerobic luminal expansion of salmonella. Cell Host Microbe 2016; 19:443–54.
https://doi.org/10.1016/j.chom.2016.03.004



27. Yale CE, Balish E. The importance of clostridia in experimental intestinal strangulation. Gastroenterology 1976; 71:793–6.
https://doi.org/10.1016/S0016-5085(76)80362-9


28. Gophna U, Konikoff T, Nielsen HB. Oscillospira and related bacteria - From metagenomic species to metabolic features. Environ Microbiol 2017; 19:835–41.
https://doi.org/10.1111/1462-2920.13658


29. Mackie Roderick I, Aminov Rustam I, Hu W, et al. Ecology of uncultivated oscillospira species in the rumen of cattle, sheep, and reindeer as assessed by microscopy and molecular approaches. Appl Environ Microbiol 2003; 69:6808–15.
https://doi.org/10.1128/AEM.69.11.6808-6815.2003



30. Chen YR, Zheng HM, Zhang GX, Chen F, Chen L, Yang Z. High Oscillospira abundance indicates constipation and low BMI in the Guangdong Gut Microbiome Project. Sci Rep 2020; 10:9364
https://doi.org/10.1038/s41598-020-66369-z



31. Magalhães VJA, Susca F, Lima FS, Branco AF, Yoon I, Santos JEP. Effect of feeding yeast culture on performance, health, and immunocompetence of dairy calves. J Dairy Sci 2008; 91:1497–509.
https://doi.org/10.3168/jds.2007-0582


32. Galvão KN, Santos JEP, Coscioni A, et al. Effect of feeding live yeast products to calves with failure of passive transfer on performance and patterns of antibiotic resistance in fecal Escherichia coli. Reprod Nutr Dev 2005; 45:427–40.
https://doi.org/10.1051/rnd:2005040


33. Ji M, Du H, Xu Y. Structural and metabolic performance of p-cresol producing microbiota in different carbon sources. Food Res Int 2020; 132:109049
https://doi.org/10.1016/j.foodres.2020.109049


34. Chaucheyras-Durand F, Walker ND, Bach A. Effects of active dry yeasts on the rumen microbial ecosystem: Past, present and future. Anim Feed Sci Technol 2008; 145:5–26.
https://doi.org/10.1016/j.anifeedsci.2007.04.019

35. Ogunade IM, Lay J, Andries K, McManus CJ, Bebe F. Effects of live yeast on differential genetic and functional attributes of rumen microbiota in beef cattle. J Anim Sci Biotechnol 2019; 10:68
https://doi.org/10.1186/s40104-019-0378-x



36. Lesmeister KE, Heinrichs AJ, Gabler MT. Effects of supplemental yeast (Saccharomyces cerevisiae) culture on rumen development, growth characteristics, and blood parameters in neonatal dairy calves. J Dairy Sci 2004; 87:1832–9.
https://doi.org/10.3168/jds.S0022-0302(04)73340-8


37. Hammes WP, Vogel RF. The genus Lactobacillus. Wood BJB, Holzapfel WH, editorsThe genera of lactic acid bacteria. Boston, MA, USA: Springer US; 1995. p. 19–54.

38. Gressley TF, Hall MB, Armentano LE. Ruminant Nutrition Symposium: Productivity, digestion, and health responses to hindgut acidosis in ruminants. J Anim Sci 2011; 89:1120–30.
https://doi.org/10.2527/jas.2010-3460


39. Sanz-Fernandez MV, Daniel J-B, Seymour DJ, et al. Targeting the hindgut to improve health and performance in cattle. Animals 2020; 10:1817
https://doi.org/10.3390/ani10101817



40. Faniyi TO, Adegbeye MJ, Elghandour MMMY, et al. Role of diverse fermentative factors towards microbial community shift in ruminants. J Appl Microbiol 2019; 127:2–11.
https://doi.org/10.1111/jam.14212


41. Jami E, White BA, Mizrahi I. Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency. PLoS One 2014; 9:e85423
https://doi.org/10.1371/journal.pone.0085423



42. Myer PR, Smith TPL, Wells JE, Kuehn LA, Freetly HC. Rumen microbiome from steers differing in feed efficiency. PLoS One 2015; 10:e0129174
https://doi.org/10.1371/journal.pone.0129174



43. Thoetkiattikul H, Mhuantong W, Laothanachareon T, et al. Comparative analysis of microbial profiles in cow rumen fed with different dietary fiber by tagged 16S rRNA gene pyrosequencing. Curr Microbiol 2013; 67:130–7.
https://doi.org/10.1007/s00284-013-0336-3


44. Biddle A, Stewart L, Blanchard J, Leschine S. Untangling the genetic basis of fibrolytic specialization by lachnospiraceae and ruminococcaceae in diverse gut communities. Diversity 2013; 5:627–40.
https://doi.org/10.3390/d5030627

45. De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA 2010; 107:14691–6.
https://doi.org/10.1073/pnas.1005963107



46. Liu J, Zhang M, Xue C, Zhu W, Mao S. Characterization and comparison of the temporal dynamics of ruminal bacterial microbiota colonizing rice straw and alfalfa hay within ruminants. J Dairy Sci 2016; 99:9668–81.
https://doi.org/10.3168/jds.2016-11398


47. Geervliet M, de Vries H, Jansen CA, et al. Effects of Escherichia coli Nissle 1917 on the porcine gut microbiota, intestinal epithelium and immune system in early life? Front Microbiol 2022; 13:842437
https://doi.org/10.3389/fmicb.2022.842437



48. Mølbak L, Klitgaard K, Jensen Tim K, Fossi M, Boye M. Identification of a novel, invasive, not-yet-cultivated treponema sp. in the large intestine of pigs by PCR amplification of the 16S rRNA gene. J Clin Microbiol 2006; 44:4537–40.
https://doi.org/10.1128/JCM.01537-06



49. Schrank K, Choi BK, Grund S, et al. Treponema brennaborense sp. nov., a novel spirochaete isolated from a dairy cow suffering from digital dermatitis. Int J Syst Bacteriol 1999; 49:43–50.
https://doi.org/10.1099/00207713-49-1-43


50. Seymour WM, Nocek JE, Siciliano-Jones J. Effects of a colostrum substitute and of dietary brewer's yeast on the health and performance of dairy calves. J Dairy Sci 1995; 78:412–20.
https://doi.org/10.3168/jds.S0022-0302(95)76650-4


51. Durso LM, Harhay GP, Smith TPL, et al. Animal-to-animal variation in fecal microbial diversity among beef cattle. Appl Environ Microbiol 2010; 76:4858–62.
https://doi.org/10.1128/AEM.00207-10



52. Kim M, Kim J, Kuehn LA, et al. Investigation of bacterial diversity in the feces of cattle fed different diets. J Anim Sci 2014; 92:683–94.
https://doi.org/10.2527/jas.2013-6841


53. Shanks OC, Kelty CA, Archibeque S, et al. Community structures of fecal bacteria in cattle from different animal feeding operations. Appl Environ Microbiol 2011; 77:2992–3001.
http://doi.org/10.1128/AEM.02988-10



54. Hernandez-Sanabria E, Goonewardene LA, Wang Z, Durunna ON, Moore SS, Guan LL. Impact of feed efficiency and diet on adaptive variations in the bacterial community in the rumen fluid of cattle. Appl Environ Microbiol 2012; 78:1203–14.
https://doi.org/10.1128/AEM.05114-11



55. Moya A, Ferrer M. Functional redundancy-induced stability of gut microbiota subjected to disturbance. Trends Microbiol 2016; 24:402–13.
https://doi.org/10.1016/j.tim.2016.02.002


56. Groves JT, Kuriyan J. Molecular mechanisms in signal transduction at the membrane. Nat Struct Mol Biol 2010; 17:659–65.
https://doi.org/10.1038/nsmb.1844



- TOOLS






 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Supplement1
Supplement1 Print
Print





