 |
 |
| Anim Biosci > Volume 33(9); 2020 > Article |
|
Abstract
Objective
Methods
Results
ACKNOWLEDGMENTS
Figure 1
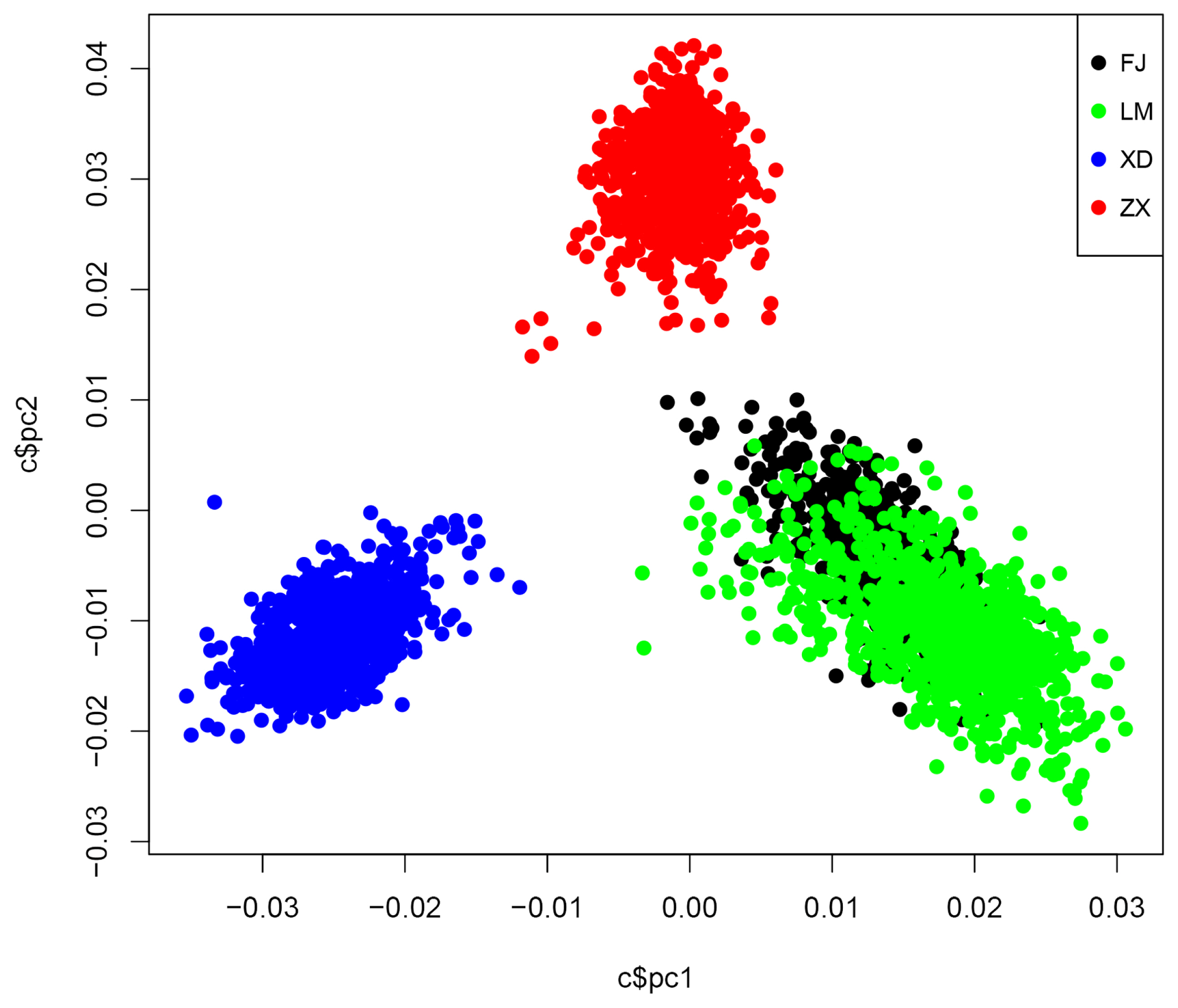
Figure 2
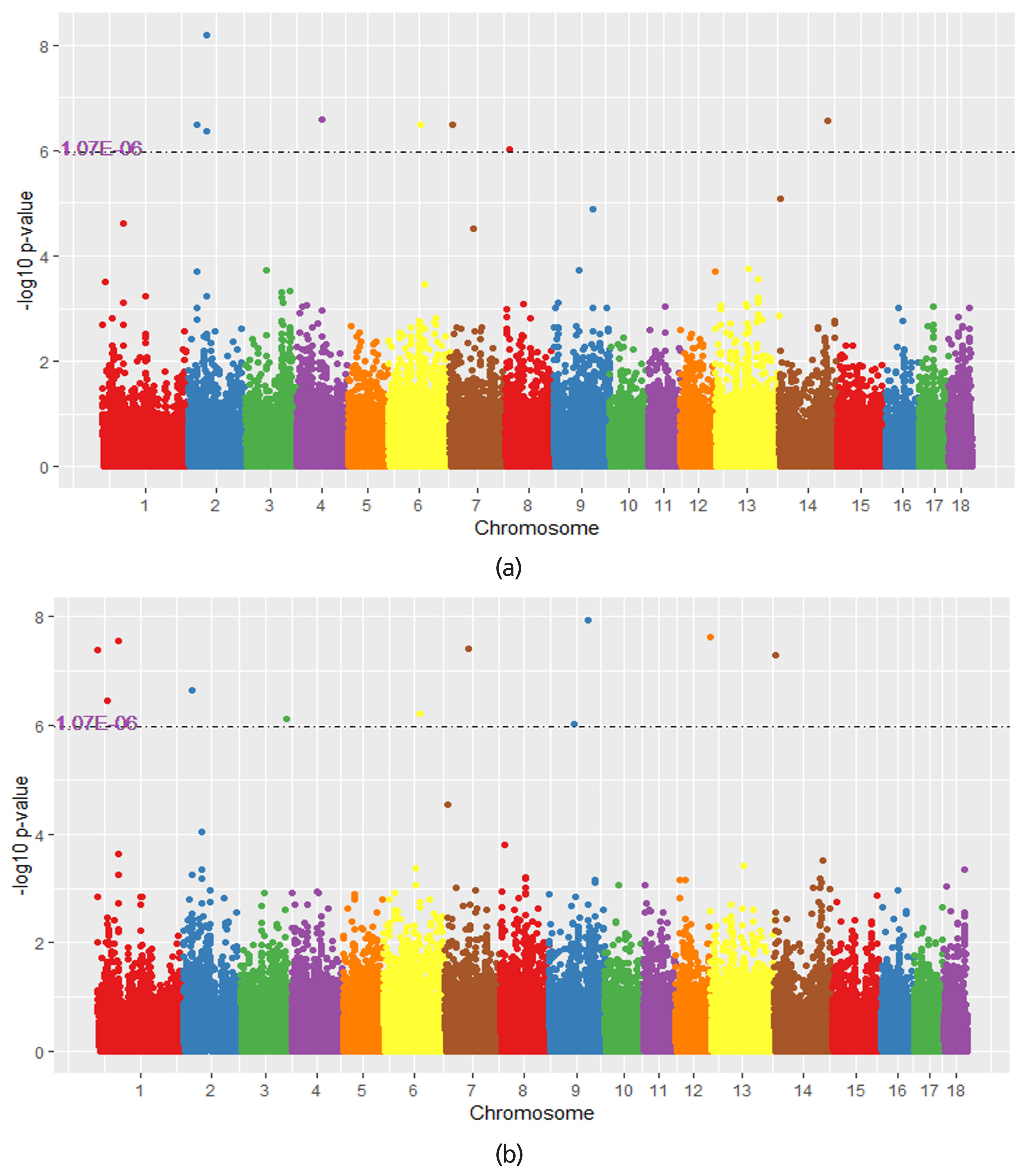
Figure 3

Table 1
| Trait | Population1) | Source | N | Min | Max | Mean | SD |
|---|---|---|---|---|---|---|---|
| NBA | LM | American line2) | 931 | 5.46 | 12.78 | 9.30 | 2.112 |
| FJ | 545 | 6.60 | 14.38 | 10.70 | 2.295 | ||
| XD | British line | 874 | 7.54 | 12.28 | 10.38 | 1.733 | |
| ZX | American line3) | 771 | 7.18 | 13.19 | 9.74 | 1.914 | |
| TNB | LM | American line2) | 931 | 6.15 | 14.38 | 10.03 | 2.226 |
| FJ | 545 | 6.89 | 16.75 | 11.73 | 2.489 | ||
| XD | British line | 874 | 8.06 | 13.59 | 10.88 | 1.810 | |
| ZX | American line3) | 771 | 7.62 | 15.00 | 11.04 | 2.155 |
Table 2
| SSC | SNP name | Location (bp) | Populations1) | p(NBA)-value2) | p(TNB)-value3) | Associated gene4) | Distance5) (bp) | Gene function |
|---|---|---|---|---|---|---|---|---|
| 17 | ALGA0094112 | 27,884,015 | XD | 1.160E-05 | 3.770E-05 | RIN2 | In | Signal transduction |
| 1 | WU_10.2_1_11153176 | 9,032,844 | XD | 1.686E-05 | 3.314E-05 | SYNJ2 | In | Inositol phosphate dephosphorylation |
| 6 | MARC0081527 | 80,617,330 | XD | 1.859E-05 | 1.780E-05 | C1QB | Down | Complement activation, classical pathway |
| 4 | WU_10.2_4_80076056 | 73,428,123 | XD | 2.188E-05 | 1.336E-05 | CA8 | Down | Phosphatidylinositol-mediated signaling |
| 2 | ALGA0113046 | 12,842,137 | XD | 2.132E-05 | 1.346E-05 | OR10Q1 | Up | G protein-coupled receptor signaling pathway |
| 5 | H3GA0015463 | 9,863,427 | XD | 3.220E-05 | 1.760E-05 | PICK1 | In | Intracellular protein transport |
| 4 | WU_10.2_4_80076056 | 73,428,123 | FJ | 1.807E-05 | 2.313E-05 | CA8 | Down | Phosphatidylinositol-mediated signaling |
| 12 | WU_10.2_12_17971455 | 41,032 | ZX | 1.080E-05 | 7.546E-04 | ZNF750 | Down | |
| 7 | WU_10.2_7_130172562 | 537,474 | ZX | 1.675E-05 | 1.040E-05 | FOXQ1 | Up | Cell differentiation |
| 1 | WU_10.2_1_11153176 | 9,032,844 | ZX | 6.517E-05 | 1.110E-05 | SYNJ2 | In | Inositol phosphate dephosphorylation |
| 7 | H3GA0021245 | 38,823,945 | ZX | 2.478E-05 | 1.125E-05 | VEGFA | Down | Regulation of signaling receptor activity |
| 1 | MARC0022141 | 0 | ZX | 1.172E-04 | 1.644E-05 | NA | NA | NA |
| 1 | DRGA0000439 | 30,456,989 | ZX | 1.918E-04 | 1.674E-05 | EYA4 | In | Protein dephosphorylation |
SSC, Sus scrofa chromosome; SNP, single nucleotide polymorphism; NBA, number of piglets born alive; TNB, total number of piglets born; RIN2, Ras and Rab interactor 2; SYNJ2, synaptojanin 2; C1QB, complement C1q B chain; CA, carbonic anhydrase 8; OR10Q1, olfactory receptor family 10 subfamily Q member 1; PICK1, protein interacting with PRKCA 1; ZNF750, zinc finger protein 750; FOXQ1, forkhead box Q1; VEGFA, vascular endothelial growth factor A; EYA4, EYA transcriptional coactivator and phosphatase 4; NA, not available; GWAS, genome-wide association study.
3) p(TNB)–value = p-value from sing population GWAS for TNB. The bold data in this column represent the SNP at suggestive genome-wide significant level.
Table 3
| SNP name | SSC | Location (bp) | p(meta-TNB)–value1) | p(meta-NBA)–value2) | Associated gene3) | Distance4) (bp) | Gene function |
|---|---|---|---|---|---|---|---|
| ALGA0012964 | 2 | 32,799,355 | 9.23E-05 | 6.41E-09 | LIN7C | In | Morphogenesis of an epithelial sheet |
| ALGA0054421 | 9 | 104,625,702 | 1.17E-08 | 1.29E-05 | LHFPL3 | In | Self reported educational attainment |
| ASGA0037579 | 8 | 5,703,865 | 0.000156302 | 9.82E-07 | MSX1 | Down | In utero embryonic development |
| ASGA0082366 | 9 | 47,037,744 | 9.84E-07 | 0.000189917 | NECTIN1 | Down | Lens morphogenesis in camera-type eye |
| ALGA0113046 | 2 | 12,842,137 | 2.29E-07 | 0.000193931 | OR10Q1 | Up | G protein-coupled receptor signaling pathway |
| ASGA0104976 | 12 | 116,492,539 | 2.44E-08 | 0.000203328 | NA | ||
| DRGA0000439 | 1 | 30,456,989 | 2.86E-08 | 2.37E-05 | EYA4 | In | Protein dephosphorylation |
| H3GA0042513 | 14 | 126,349,106 | 0.000299386 | 2.80E-07 | GFRA1 | In | Nervous system development |
| WU_10.2_7_130172562 | 7 | 537,474 | 2.88E-05 | 3.27E-07 | FOXQ1 | Up | Cell differentiation |
| H3GA0021245 | 7 | 38,823,945 | 3.95E-08 | 3.11E-05 | VEGFA | Down | Regulation of signaling receptor activity |
| MARC0022141 | 1 | 0 | 4.11E-08 | 0.000318796 | NA | NA | NA |
| WU_10.2_4_80076056 | 4 | 73,428,123 | 0.00116 | 2.62E-07 | CA8 | Down | Phosphatidylinositol-mediated signaling |
| WU_10.2_1_11153176 | 1 | 9,032,844 | 3.51E-07 | 0.001559 | SYNJ2 | In | Inositol phosphate dephosphorylation |
| WU_10.2_3_129122235 | 3 | 120,870,358 | 7.72E-07 | 0.060399 | FAM49A | Up | NA |
| MARC0081527 | 6 | 80,617,330 | 0.000432722 | 3.20E-07 | C1QB | Down | Complement activation, classical pathway |
| WU_10.2_14_389214 | 14 | 217,583 | 5.23 E-08 | 8.22E-06 | SPIN1 | Up | Wnt signaling pathway |
| WU_10.2_2_12776809 | 2 | 13,143,791 | 0.000571848 | 3.33E-07 | CTNND1 | Up | Negative regulation of canonical Wnt signaling pathway |
| WU_10.2_6_85867859 | 6 | 92,797,244 | 6.21E-07 | 0.000352248 | GRIK3 | Up | Glutamate receptor signaling pathway |
| ALGA0012962 | 2 | 32,757,436 | 0.00064956 | 4.23E-07 | LIN7C | In | Morphogenesis of an epithelial sheet |
SNP, single nucleotide polymorphism; SSC, Sus scrofa chromosome; TNB, total number of piglets born; NBA, number of piglets born alive; LIN7C, lin-7 homolog C; LHFPL3, LHFPL tetraspan submily member 3; MSX1, msh homeobox 1; NECTIN1, nectin cell adhesion molecule 1; OR10Q1, olfactory receptor family 10 subfamily Q member 1; EYA4, EYA transcriptional coactivator and phosphatase 4; GFRA1, GDNF family receptor alpha 1; FOXQ1, forkhead box Q1; VEGFA, vascular endothelial growth factor A; CA8, carbonic anhydrase 8; SYNJ2, synaptojanin 2; FAM49A, family with sequence similarity 49 member A; C1QB, complement C1q B chain; SPIN1, spindlin 1; CTNND1, catenin delta 1; GRIK3, glutamate ionotropic receptor kainate type subunit 3; VA, not available.
1) p(meta-TNB)–value = p-value from the meta analysis. The bold data in this column represent the significant SNP at genome-wide significant level; otherwise at the chromosome-wide significant level.
2) p(meta-NBA)–value = p-value from the meta analysis. The bold data in this column represent the significant level.
Table 4
| SNP name | SSC | Location (bp) | p(meta- LM)–value1) | p(meta- FJ)–value2) | p(meta- XD)–value3) | p(meta- ZX)–value4) | Associated gene5) | Distance6) (bp) | Gene function |
|---|---|---|---|---|---|---|---|---|---|
| ALGA0012964 | 2 | 32,799,355 | 1.04E-06 | 2.68E-06 | 1.08E-05 | 2.65E-08 | LIN7C | In | Morphogenesis of an epithelial sheet |
| ALGA0054421 | 9 | 104,625,702 | 1.24E-07 | 6.30E-08 | 4.47E-08 | 1.14E-05 | LHFPL3 | In | Self reported educational attainment |
| ASGA0037579 | 8 | 5,703,865 | 3.01E-06 | 3.75E-08 | 1.53E-06 | 1.37E-07 | MSX1 | Down | In utero embryonic development |
| DRGA0000439 | 1 | 30,456,989 | 3.74E-07 | 3.93E-07 | 3.74E-07 | 6.60E-08 | EYA4 | In | Protein dephosphorylation |
| H3GA0021245 | 7 | 38,823,945 | 1.37E-07 | 7.92E-06 | 3.82E-07 | 6.41E-09 | VEGFA | Down | Regulation of signaling receptor activity |
| H3GA0042513 | 14 | 126,349,106 | 2.31E-07 | 2.35E-06 | 4.84E-07 | 1.07E-05 | GFRA1 | In | Nervous system development |
| MARC0081527 | 6 | 80,617,330 | 3.34E-08 | 1.45E-07 | 7.55E-09 | 7.35E-06 | C1QB | Down | Complement activation, classical pathway |
| WU_10.2_14_389214 | 14 | 217,583 | 4.59E-06 | 3.02E-08 | 1.06E-05 | 1.64E-06 | SPIN1 | Up | Wnt signaling pathway |
| WU_10.2_2_12776809 | 2 | 13,143,791 | 1.57E-08 | 2.52E-06 | 1.99E-06 | 3.78E-06 | CTNND1 | Up | Negative regulation of canonical Wnt signaling pathway |
| WU_10.2_6_85867859 | 6 | 92,797,244 | 5.78E-07 | 2.26E-06 | 4.41E-06 | 1.53E-06 | GRIK3 | Up | Glutamate receptor signaling pathway |
| WU_10.2_7_130172562 | 7 | 537,474 | 1.96E-07 | 4.10E-06 | 1.88E-06 | 4.09E-09 | FOXQ1 | Up | Cell differentiation |
SNP, single nucleotide polymorphism; LIN7C, lin-7 homolog C; LHFPL3, LHFPL tetraspan submily member 3; MSX1, msh homeobox 1; EYA4, EYA transcriptional coactivator and phosphatase 4; VEGFA, vascular endothelial growth factor A; GFRA1, GDNF family receptor alpha 1; C1QB, complement C1q B chain; SPIN1, spindlin 1; CTNND1, catenin delta 1; GRIK3, glutamate ionotropic receptor kainate type subunit 3; FOXQ1, forkhead box Q1.
1)–4) p(meta-XX)–value = p-value from the multi-traits meta analysis for four populations LM, FJ, XD, and ZX. The bold data in this column represent the significant SNP at genome-wide significant level; otherwise at the chromosome-wide significant level.






 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Supplement
Supplement Print
Print





