 |
 |
| Anim Biosci > Volume 32(7); 2019 > Article |
|
Abstract
Objective
Mutations in low-density lipoprotein receptor (LDLR), which encodes a critical protein for cholesterol homeostasis and lipid metabolism in mammals, are involved in cardiometabolic diseases, such as familial hypercholesterolemia in pigs. Whereas microRNAs (miRNAs) can control LDLR regulation, their involvement in circulating cholesterol and lipid levels with respect to cardiometabolic diseases in pigs is unclear. We aimed to identify and analyze LDLR as a potential target gene of SSC-miR-20a.
Methods
Bioinformatic analysis predicted that porcine LDLR is a target of SSC-miR-20a. Wild-type and mutant LDLR 3′-untranslated region (UTR) fragments were generated by polymerase chain reaction (PCR) and cloned into the pGL3-Control vector to construct pGL3 Control LDLR wild-3′-UTR and pGL3 Control LDLR mutant-3′-UTR recombinant plasmids, respectively. An miR-20a expression plasmid was constructed by inserting the porcine pre-miR-20a-coding sequence between the HindIII and BamHI sites in pMR-mCherry, and constructs were confirmed by sequencing. HEK293T cells were co-transfected with the miR-20a expression or pMR-mCherry control plasmids and constructs harboring the corresponding 3′-UTR, and relative luciferase activity was determined. The relative expression levels of miR-20a and LDLR mRNA and their correlation in terms of expression levels in porcine liver tissue were analyzed using reverse-transcription quantitative PCR.
Results
Gel electrophoresis and sequencing showed that target gene fragments were successfully cloned, and the three recombinant vectors were successfully constructed. Compared to pMR-mCherry, the miR-20a expression vector significantly inhibited wild-type LDLR-3′-UTR-driven (p<0.01), but not mutant LDLR-3′-UTR-driven (p>0.05), luciferase reporter activity. Further, miR-20a and LDLR were expressed at relatively high levels in porcine liver tissues. Pearson correlation analysis revealed that porcine liver miR-20a and LDLR levels were significantly negatively correlated (r = −0.656, p<0.05).
Serum lipids, including total cholesterol, low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol, and triglycerides, are important clinical diagnostic indices of cardiovascular diseases [1]. Despite progress in the treatment of abnormal levels of cholesterol and other lipids, cardiometabolic diseases represent prominent causes of human morbidity and mortality [2]. Pigs are widely used in biomedical research [3,4]. As their heart size, blood supply, coronary system function, and aorta features are comparable to those in humans, the pig is an important model for cardiovascular disease research [5]. Several candidate genes for porcine serum lipids, such as low-density lipoprotein receptor (LDLR), apolipoprotein B, and low density lipoprotein receptor adapter protein 1 (LDLRAP1), have been reported to date [6]. The LDLR is a cell membrane glycoprotein that plays a critical role in cholesterol homeostasis and lipid metabolism in mammals [7]. Further, LDLR gene mutations play a major role in the progression of cardiometabolic diseases [8]. In pigs, loss-of-function mutations in this gene result in the accumulation of LDL-C in circulation, leading to familial hypercholesterolemia [9], a severe disease characterized by premature death due to accelerated cardiovascular disease. LDL-C uptake by the LDLR in the liver is of clinical importance as LDLR-dependent hepatic clearance of circulating LDL-C has a prominent role in preventing atherosclerosis and cardiovascular disease [10]. The recent finding that manipulating miRNA expression might reduce circulating LDL-C could lead to a new option for the treatment of dyslipidemias [11].
miRNAs, comprising a class of short (20 to 24 nt) regulatory RNAs that modulate mRNA translation and turnover, have recently emerged as crucial regulators of mammalian cholesterol and lipid homeostasis [12]. Several miRNAs have been found to control aspects of cholesterol and lipid homeostasis at the cellular level and in vivo [13]. Specifically, recent work uncovered a role for miRNAs in directly controlling the LDLR activity in vitro and in vivo by post-transcriptionally regulating the LDLR gene [14]. This miRNA-centered regulatory network continues to grow rapidly and will undoubtedly expand to include other miRNAs that regulate LDL-C levels. Future studies using non-human primates will likely help determine the relative contribution of each miRNA in controlling LDLR activity and cholesterol homeostasis [15]. However, the potential contribution of miRNAs to dysregulated circulating cholesterol and lipids and associated cardiometabolic diseases in pigs remains unclear.
Because of the physiological and clinical importance of the LDLR in regulating circulating LDL-C, and as porcine LDLR expression was not previously shown to be under miRNA control, we conducted bioinformatic analysis to predict miRNAs that could bind the porcine LDLR 3′-untranslated region (UTR). We found that Sus scrofa (SSC)-miR-20a could directly bind LDLR mRNA with low binding free energy. Next, we constructed an SSC-LDLR 3′-UTR dual luciferase reporter vector, a vector encoding the 3′-UTR mutated at the miR-20a-binding site, and an SSC-miRNA-20a expression plasmid. A dual luciferase assay system was used to detect relative luciferase activities. Additionally, we assessed the correlation between miR-20a and LDLR expression levels in porcine liver tissues. This study aimed to provide a theoretical basis for elucidating the direct relationship between SSC-miR-20a and LDLR, as well as the possible mechanism through which miR-20a and LDLR prevent dyslipidemias and cardiovascular diseases.
All animal experiments were conducted according to the Regulations and Guidelines for Experimental Animals established by the Ministry of Science and Technology (Beijing, China, revised in 2004) and approved by the Institutional Animal Care and Use Committee of Anhui Agricultural University (Permit number: SYXK 2016-007).
Human embryonic kidney (HEK) 293T cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (Invitrogen Inc., Carlsbad, CA, USA) containing 10% fetal bovine serum. The cells were maintained at 37°C in the presence of 5% CO2. Liver, heart, spleen, lung, and kidney tissues from 12 Large White castrated boars of approximately 100 kg were collected. Tissues were frozen in liquid nitrogen immediately after resection and stored at −80°C until use.
We retrieved miRNAs from the miRBase database (http://www.mirbase.org/) that might associate with the SSC-LDLR 3′-UTR using miRanda (http://www.microrna.org), miRDB (http://www.mirdb.org/mirdb/policy.html), and Targetscan (http://www.Targetscan.org) software. Based on predictive criteria, including having good complementarity with targeted sequences, being bound to targeted sequences with low free energy of binding, and being conserved among species, miR-20a was selected as a candidate miRNA for follow-up studies.
Total RNA was extracted from porcine liver tissues using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer’s protocol. RNA concentrations and purity were measured with an ND 2000 spectrophotometer (NanoDrop; Thermo Fisher Scientific, Inc., Wilmington, DE, USA).
For the quantitative analysis of miR-20a expression, 1 μg of total RNA from each sample was reverse-transcribed into cDNA using the NCode EXPRESS SYBR GreenER miRNA qPCR Kit (Invitrogen, USA) at 37°C for 1 h and 85°C for 5 s, in a 20-μL reaction mixture, according to the manufacturer’s instructions. With reference to miRBase 21.0 (http://www.mirbase.org/), we designed miRNA-specific forward primers by poly (A)-tailing miR-20a; the universal reverse primer was provided in the Invitrogen kit. Porcine U6 small nuclear RNA (U6) was used as an endogenous control gene.
For quantitative analysis of LDLR mRNA expression, 1 μg of total RNA from each sample was reverse-transcribed into cDNA using the Prime Script RT Kit (TaKaRa, Tokyo, Japan) at 37°C for 15 min and 85°C for 5 s in a 10-μL reaction mixture, according to the manufacturer’s instructions. The β-actin gene was used as an endogenous control gene.
All primers used are listed in Table 1. Quantitative polymerase chain reaction (qPCR) was performed in triplicate using a CFX96 Touch real-time PCR detection system (Bio-Rad, Hercules, CA, USA). Thermal cycling conditions were as follows: 94°C for 30 s, followed by 40 cycles at 94°C for 5 s, and 60°C for 30 s. The relative expression of miR 20a and LDLR mRNA was determined using the 2−ΔΔCt method [16].
For the functional analysis of miR-20a, a partial segment of the LDLR mRNA 3′-UTR containing the miR-20a-binding sequence and a mutated segment of the LDLR mRNA 3′-UTR in which the miR-20a-binding sequence AAGCACTG was converted to AACGACTG, were PCR amplified from DNA prepared from porcine liver tissues. Based on the porcine LDLR mRNA 3′-UTR sequence (NCBI: NM_001206354.2), primer sequences were designed (Table 2). The expected length of both amplicons was 688 bp. The PCR products were subcloned into the XbaI site downstream of the stop codon in the PGL3-Control firefly luciferase reporter vector (Promega, Madison, WI, USA; the vector backbone is shown in Figure 1). Recombinants harboring the desired PCR products were confirmed by DNA sequencing. The recombinant plasmids were termed “WT” (pGL3 Control LDLR wild-3′-UTR) and “MT” (pGL3 Control LDLR mutant -3′-UTR).
Pre-miR-20a coding sequences were PCR-amplified from cDNA prepared from porcine liver tissues. Primer 5.0 was used to design PCR primers based on the sequence of porcine miR-20a (NCBI: MIMAT0002129). Primer sequences are shown in Table 2. The expected length of the amplicon was 170 bp. MiR-20a expression plasmids were constructed by inserting a DNA fragment containing the pre-miRNA coding sequence between the HindIII and BamHI (Fermentas, Vilnius, Lithuania) sites of pMR-mCherry (Clontech, Palo Alto, CA, USA; the vector backbone is shown in Figure 2). The constructs containing the pre-miRNA coding sequences were confirmed by sequencing and termed “miR-20a expression vectors”.
Plasmids were transfected using Lipofectamine 2000 (Invitrogen, USA) according to the supplier’s protocol. For luciferase assays, HEK293T cells at 70% confluence were transferred into 24-well plates with miR-20a expression plasmids or the pMR-mCherry control plasmid, as well as the firefly luciferase constructs containing the corresponding 3′-UTR. Luciferase activity was measured 24 h post-transfection using the Dual-Luciferase Reporter 1000 System (Promega, USA), based on the manufacturer’s protocol. Briefly, cells were lysed with passive lysis buffer at room temperature for 15 min. Luciferase assay buffer II was then added, and firefly luciferase activity was immediately determined using a Fluoroskan Ascent FL microplate reader (Thermo Scientific, Waltham, MA, USA). Next, Stop & Glo Buffers with Stop & Glo substrates were added and mixed briefly. Renilla luciferase activity was immediately determined and firefly luciferase activity was normalized to Renilla luciferase activity to account for variations in transfection efficiency. All reactions were performed in triplicate.
Data are expressed as the mean±standard error of the mean. A Student’s t-test was applied to compare two groups and one-way analysis of variance was performed for multiple groups; the association between liver miR-20a and LDLR mRNA expression levels was analyzed by calculating the Pearson’s correlation coefficient. Analyses were conducted with SPSS software (version 19.0; IBM, Armonk, NY, USA). Less than 0.05 p value was considered to indicate a statistically significant difference and less than 0.01 p value was considered to indicate an extremely statistically significant difference.
Using miRanda, TargetScan, and miRDB programs, LDLR was predicted as a potential target of miR-20a (Figure 3). As shown in Figure 3a, the SSC-miR-20a and LDLR 3′-UTR sequences were determined to have good complementarity and to bind each other with low binding free energy (approximately −23.0 kcal/mol). As shown in Figure 3b, one potential target site for SSC-miR-20a was identified in the LDLR 3′-UTR. These results suggested that LDLR is a target of SSC-miR-20a.
A partial segment of the LDLR mRNA 3′-UTR containing the miR-20a-binding sequence and a mutated segment of the LDLR mRNA 3′-UTR in which the miR-20a-binding sequence AAGCACTG was converted to AACGACTG, were PCR amplified from DNA prepared from porcine liver tissues. Gel electrophoresis results showed that the PCR products were specific and of the expected size (Figure 4).
Sequencing results of the pGL3-Control-LDLR-wild-3′-UTR vector are shown in Figure 5a; the cloned fragment had 100% sequence identity with the corresponding region of the porcine LDLR gene listed in GenBank. Sequencing results of the pGL3-Control-LDLR-mutant-3′-UTR vector are shown in Figure 5b; the AAGCACT sequence was successfully mutated to AACG ACT, without changes to other bases. These results confirmed that LDLR-wild-3′-UTR and LDLR-mutant-3′-UTR were successfully cloned into the dual luciferase reporter vectors. Sequencing results of the miR-20a expression vector are shown in Figure 5c; the sequence of the miR-20a fragment was in good agreement with the designed pre-miR-20a coding sequence, indicating that the miR-20a expression vector was successfully constructed.
Firefly luciferase activities in HEK293T cells transfected with the recombinant plasmids are shown in Figure 6. Compared to the pMR-mCherry control, the miR-20a expression vector significantly inhibited wild-type LDLR-3′-UTR activity (p< 0.01). However, the miR-20a expression vector had no effect on luciferase activity from the LDLR mRNA 3′-UTR reporter containing the mutated miR-20a-binding site. These results indicated that miR-20a targets LDLR via the miR-20a-binding site located in the LDLR mRNA 3′-UTR and inhibits its transcriptional activity, which supports the contention that LDLR is directly regulated by miR-20a.
Relative expression levels of miR-20a and LDLR mRNA in liver, heart, spleen, lung, and kidney tissues of 12 Large White pigs were analyzed using reverse transcription (RT)-qPCR. As shown in Figure 7, SSC-miR-20a and LDLR were relatively highly expressed in porcine liver tissues. To test whether miR-20a targets LDLR mRNA in porcine liver tissues, we analyzed the correlation between these two markers. Pearson correlation analysis revealed that in porcine livers, miR-20a and LDLR expression levels were significantly negatively correlated (r = −0.656, p<0.05).
Atherosclerosis is primarily caused by an imbalance of blood lipids, and in particular, the accumulation of LDL-C particles plays a significant role in disease progression [17]. The hepatic LDLR pathway is essential for clearing circulating LDL-C. Whereas the transcriptional regulation of LDLR is well characterized, the post-transcriptional mechanisms that govern its expression are only beginning to emerge [18]. The contribution of miRNAs to the post-transcriptional regulation of LDLR expression has recently been demonstrated. Notably, miR-27a/b, miR-185, miR-199a, miR-148a, miR-128-1, miR-130b, and miR-301 were shown to directly target the 3′-UTR of LDLR to modulate its expression in human and mouse hepatic cells [19–21]. Recent evidence also suggests that miR-20a contributes to atherosclerosis progression by targeting genes encoding ATP-binding cassette transporter A1 and phosphatase and tensin homolog to regulate their expression [22,23].
In the present study, bioinformatic analysis predicted that SSC-miR-20a, which shares 100% and 94.4% nucleotide similarity with human and mouse orthologs, respectively, directly binds porcine LDLR mRNA with low free energy of binding. A luciferase-reporter assay confirmed that SSC-miR-20a binds a LDLR 3′-UTR construct. In addition, SSC-miR-20a and LDLR were highly expressed in the porcine liver, and liver miR-20a and LDLR expression levels were significantly negatively correlated. Together, our findings indicated that SSC-miR-20a might act as a negative regulator of porcine LDLR expression and protein activity.
The liver is the main site of circulating LDL-C clearance, and LDLR plays a crucial role in hepatic LDL-C uptake [10]. The high expression of SSC-miR-20a and LDLR in the porcine liver suggested that miR-20a is a novel important regulator of hepatic LDL-C clearance, and functions through the direct regulation of LDLR expression. Bioinformatics predictions and experimental approaches have previously indicated that a single miR can target more than 100 mRNAs, and similarly, one gene can be regulated by multiple miRs [24]. Given the diversity of miRNAs and their targets, the mechanism and function of miR-specific regulation of mRNAs are complicated. However, highly expressed miRs generally have an inhibitory effect on target mRNA expression. In our model (12 Large White castrated boars with body weights of approximately 100 kg), porcine liver miR-20a was found to be negatively correlated with LDLR mRNA expression levels; this warrants further, larger studies to confirm the effect of miR-20a on inhibiting LDLR expression.
In conclusion, luciferase activity and RT-qPCR assays suggested that LDLR is a target of SSC-miR-20a. These results, suggesting that SSC-miR-20a can bind LDLR, indicate that further study is required to fully elucidate the role of miR-20a in the pathogenesis and progression of cardiovascular diseases.
ACKNOWLEDGMENTS
This work was supported by grants from the Natural Science Foundation of Anhui Province (1508085QC52), the key subject of study and research of Anhui province (1704a07020085, 1704a07020090), the National Natural Science Foundation of China (31572377), The planning subject of ‘the twelfth five-year-plan’ in national science and technology for the rural development in China (2015BAD03B01), Anhui Provincial Science and Technology Major Project (17030701008, 17030 701061), the Open Fund of Anhui Province Key Laboratory of Local Livestock and Poultry (AKLGRCB2017003), the Anhui Provincial Modern Industrial Technology System of Swine, and the Open project supported by State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences (GREKF15-03). We would like to thank Editage [www.editage.cn] for English language editing.
Figure 3
Target prediction results of porcine LDLR 3′-UTR binding. (a) Comparison of base-pair stem-and-loop structure between mature SSC-miR-20a and LDLR 3′-UTR. (b) A potential target site for SSC-miR-20a was found to be located in the 3′-UTR of the LDLR gene. LDLR, low-density lipoprotein receptor; UTR, untranslated region.
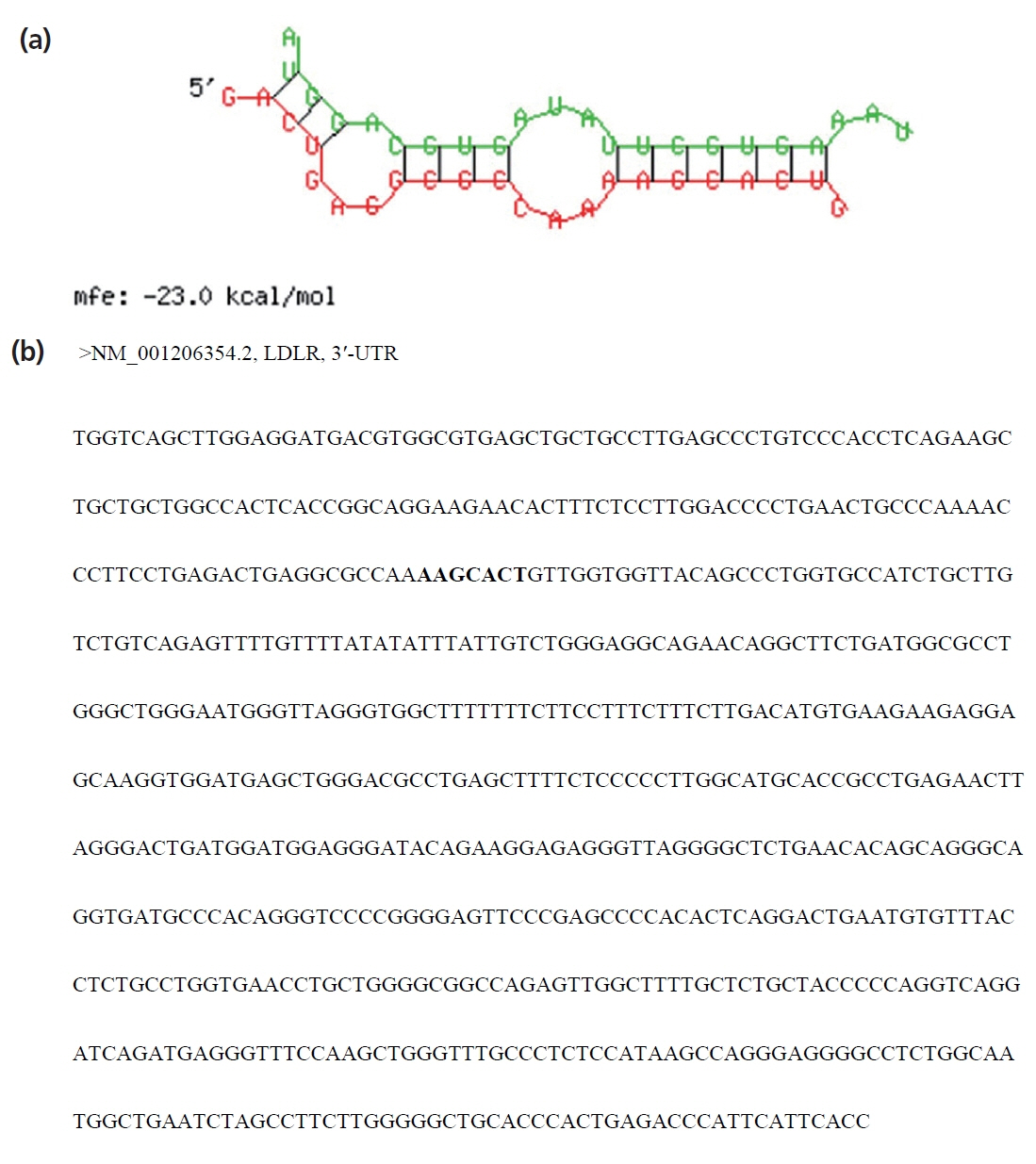
Figure 4
Agarose gel electrophoresis analysis of PCR products. M: DNA marker; lane 1: PCR product of miR-20a precursor; lane 2: PCR product of LDLR mRNA 3′-UTR sequence; lane 3: PCR product of LDLR mRNA 3′-UTR mutant sequence. PCR, polymerase chain reaction; LDLR, low-density lipoprotein receptor; UTR, untranslated region.
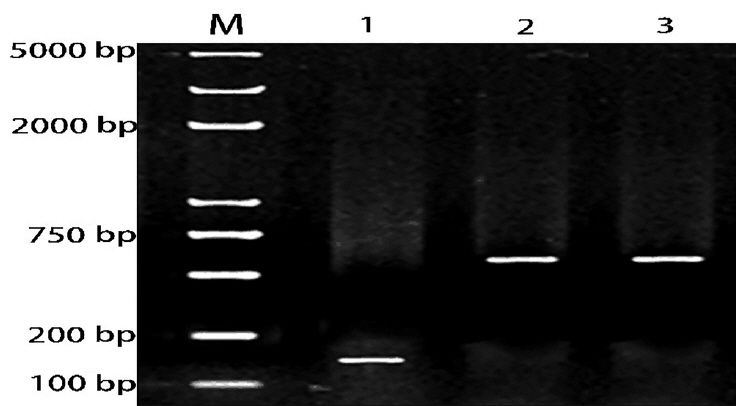
Figure 5
Sequencing results of recombinant plasmids. (a) Sequencing results of dual luciferase reporter vector containing the LDLR 3′-UTR; (b) sequencing results of dual luciferase reporter vector containing the LDLR 3′-UTR with a target sequence mutation; (c) sequencing results of miR-20a expression vector. LDLR, low-density lipoprotein receptor; UTR, untranslated region.
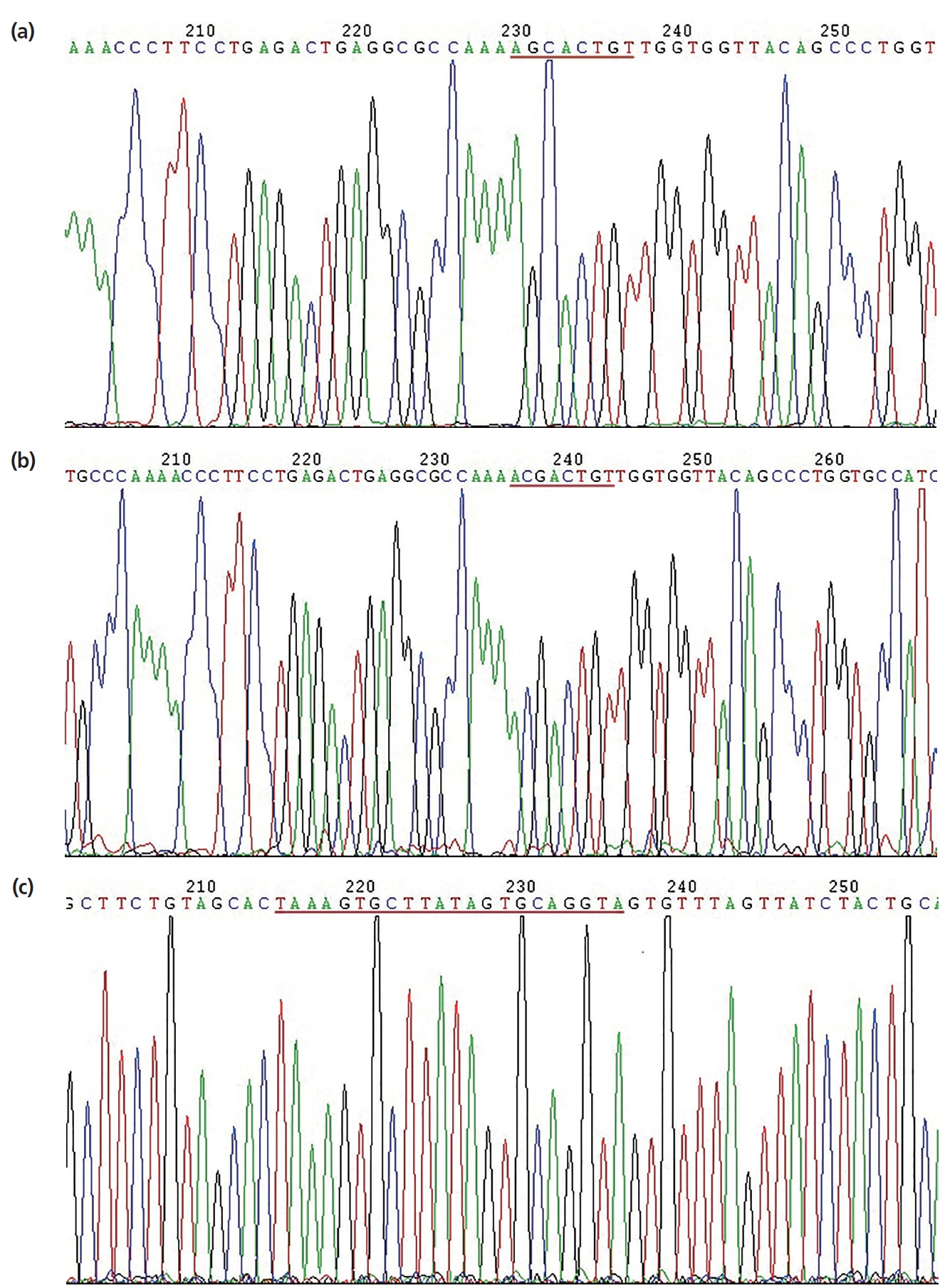
Figure 6
Luciferase activity in HEK293T cells transfected with reporter vectors containing LDLR mRNA 3′-UTR variants and an miR-20a expression vector. HEK293T, human embryonic kidney 293T; LDLR, low-density lipoprotein receptor; UTR, untranslated region; WT, wild-type LDLR 3′-UTR; MT, LDLR 3′-UTR with a target sequence mutation in miR-20a-binding site. Error bars represent the standard error of three independent experiments. *** p<0.01.
Figure 7
Expression of miR-20a and LDLR in different tissues of Large White pigs (n = 12). Expression was determined by RT-qPCR. LDLR, low-density lipoprotein receptor; RT-qPCR, reverse transcription quantitative polymerase chain reaction.
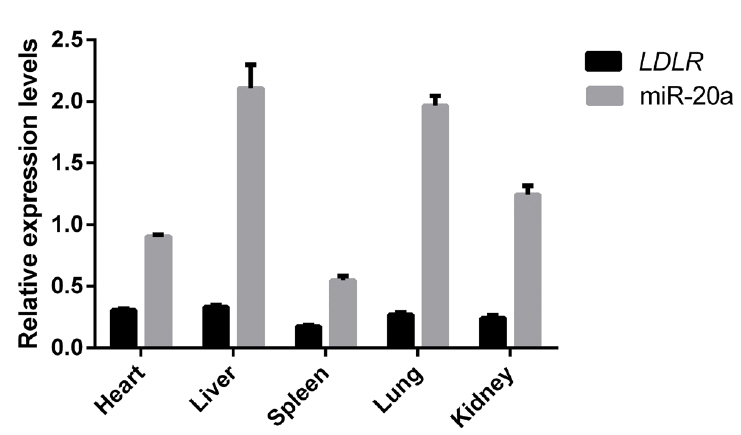
Table 1
Sequences of primers used for reverse transcription quantitative polymerase chain reaction
Table 2
Sequences of primers used to amplify gene fragments and miRNAs
REFERENCES
1. Bell DA, Bender R, Hooper AJ, et al. Impact of interpretative commenting on lipid profiles in people at high risk of familial hypercholesterolaemia. Clin Chim Acta 2013; 422:21–5. https://doi.org/10.1016/j.cca.2013.03.027


2. Rodríguez-Pérez JM, Posadas-Sánchez R, Blachman-Braun R, et al. HHIPL-1 (rs2895811) gene polymorphism is associated with cardiovascular risk factors and cardiometabolic parameters in Mexicans patients with myocardial infarction. Gene 2018; 663:34–40. https://doi.org/10.1016/j.gene.2018.04.030


3. Vilahur G, Padro T, Badimon L. Atherosclerosis and thrombosis: insights from large animal models. J Biomed Biotechnol 2011; 2011:Article ID 907575. http://dx.doi.org/10.1155/2011/907575


4. Wang Y, Du Y, Shen B, et al. Efficient generation of gene-modified pigs via injection of zygote with Cas9/sgRNA. Sci Rep 2015; 5:8256http://dx.doi.org/10.1038/srep08256



5. Chen F, Wang Y, Yuan Y, et al. Generation of B cell-deficient pigs by highly efficient CRISPR/Cas9-mediated gene targeting. J Genet Genomics 2015; 42:437–44. https://doi.org/10.1016/j.jgg.2015.05.002


6. Zeng Z, Chen R, Liu C, Yang H, Chen C, Huang L. Evaluation of the causality of the low-density lipoprotein receptor gene (LDLR) for serum lipids in pigs. Anim Genet 2015; 45:665–73. https://doi.org/10.1111/age.12183

7. Goldstein JL, Brown MS. The LDL receptor. Arterioscler Thromb Vasc Biol 2009; 29:431–8. https://doi.org/10.1161/ATVBAHA.108.179564



8. Huang L, Hua Z, Xiao H, et al. CRISPR/Cas9-mediated ApoE−/− and LDLR−/− double gene knockout in pigs elevates serum LDL-C and TC levels. Oncotarget 2017; 8:37751–60. https://doi.org/10.18632/oncotarget.17154



9. Pena RN, Cánovas A, Varona L, et al. Nucleotide sequence and association analysis of pig apolipoprotein-B and LDL-receptor genes. Anim Biotechnol 2009; 20:110–23. https://doi.org/10.1080/10495390902892518


10. Brown MS, Goldstein JL. How LDL receptors influence cholesterol and atherosclerosis. Sci Am 1984; 251:58–66.

11. Momtazi AA, Banach M, Pirro M, Stein EA, Sahebkar A. MicroRNAs: new therapeutic targets for familial hypercholesterolemia? Clin Rev Allergy Immunol 2018; 54:224–33. https://doi.org/10.1007/s12016-017-8611-x



12. Quiat D, Olson EN. MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment. J Clin Invest 2013; 123:11–8. https://doi.org/10.1172/JCI62876



13. Goedeke L, Aranda JF, Fernándezhernando C. microRNA regulation of lipoprotein metabolism. Curr Opin Lipidol 2014; 25:282–8. https://doi.org/10.1097/MOL.0000000000000094



14. Goedeke L, Wagschal A, Fernándezhernando C, Näär AM. miRNA regulation of LDL-cholesterol metabolism. Biochim Biophys Acta 2016; 1861:2047–52. https://doi.org/10.1016/j.bbalip.2016.03.007



15. Aryal B, Singh AK, Rotllan N, Price N, Fernándezhernando C. microRNAs and lipid metabolism. Curr Opin Lipidol 2017; 28:273–80. https://doi.org/10.1097/MOL.0000000000000420



16. Zhang XD, Feng YF, Zhang X, et al. Differential expression of miR-145, miR-429 and its target genes in partial reproductive tissues of swine with high and low litter size. Ann Anim Sci 2017; 17:671–81. https://doi.org/10.1515/aoas-2016-0082


17. Soundarya PK, Nirmala P, Ashok KP, Krishna PT. Effect of Lutein in the expression of PPARα and LDLR in hypercholesterolemic male Wistar rats. Int J Basic Clin Pharmacol 2018; 7:684–90. https://doi.org/10.18203/2319-2003.ijbcp20181170

18. Goedeke L, Rotlan N, Canfran-Duque A, et al. MicroRNA-148a regulates LDL receptor and ABCA1 expression to control circulating lipoprotein levels. Nat Med 2015; 21:1280–9. https://doi.org/10.1038/nm.3949




19. Jiang H, Zhang J, Du Y, et al. microRNA-185 modulates low density lipoprotein receptor expression as a key posttranscriptional regulator. Atherosclerosis 2015; 243:523–32. https://doi.org/10.1016/j.atherosclerosis.2015.10.026


20. Alvarez ML, Khosroheidari M, Eddy E, Done SC. MicroRNA-27a decreases the level and efficiency of the ldl receptor and contributes to the dysregulation of cholesterol homeostasis. Atherosclerosis 2015; 242:595–604. https://doi.org/10.1016/j.atherosclerosis.2015.08.023



21. Wagschal A, Najafi-Shoushtari SH, Wang L, et al. Genome-wide identification of microRNAs regulating cholesterol and triglyceride homeostasis. Nat Med 2015; 21:1290–7. https://doi.org/10.1038/nm.3980




22. Wang D, Wang Y, Ma J, et al. MicroRNA-20a participates in the aerobic exercise-based prevention of coronary artery disease by targeting PTEN. Biomed Pharmacother 2017; 95:756–63. https://doi.org/10.1016/j.biopha.2017.08.086


23. Liang B, Wang X, Song X, et al. MicroRNA-20a/b regulates cholesterol efflux through post-transcriptional repression of ATP-binding cassette transporter A1. Biochim Biophys Acta Mol Cell Biol Lipids 2017; 1862:929–38. https://doi.org/10.1016/j.bbalip.2017.06.002


24. Brümmer A, Hausser J. MicroRNA binding sites in the coding region of mRNAs: extending the repertoire of post-transcriptional gene regulation. Bioessays 2014; 36:617–26. https://doi.org/10.1002/bies.201300104








 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print





