 |
 |
| Anim Biosci > Volume 31(8); 2018 > Article |
|
Abstract
Objective
A disintegrin and metallopeptidase with thrombospondin motifs type 8 (ADAMTS8) is crucial for diverse physiological processes, such as inflammation, tissue morphogenesis, and tumorigenesis. The chicken ADAMTS8 (chADAMTS8) gene was differentially expressed in the kidney following exposure to different calcium concentrations, suggesting a pathological role of this protein in metabolic diseases. We aimed to examine the molecular characteristics of chADAMTS8 and analyze the gene-expression differences in response to toll-like receptor 3 (TLR3) stimulation.
Methods
The ADAMTS8 mRNA and amino acid sequences of various species (chicken, duck, cow, mouse, rat, human, chimpanzee, pig, and horse) were retrieved from the Ensembl database and subjected to bioinformatics analyses. Reverse-transcription polymerase chain reaction (RT-PCR) and quantitative PCR (qPCR) experiments were performed with various chicken tissues and the chicken fibroblast DF-1 cell line, which was stimulated with polyinosinic-polycytidylic acid (poly[I:C]; a TLR3 ligand).
Results
The chADAMTS8 gene was predicted to contain three thrombospondin type 1 (TSP1) domains, whose amino acid sequences shared homology among the different species, whereas sequences outside the TSP1 domains (especially the amino-terminal region) were very different. Phylogenetic analysis revealed that chADAMTS8 is evolutionarily clustered in the same clade with that of the duck. chADAMTS8 mRNA was broadly expressed in chicken tissues, and the expression was significantly up-regulated in the DF-1 cells in response to poly(I:C) stimulation (p<0.05). These results showed that chADAMTS8 may be a target gene for TLR3 signaling.
Conclusion
In this report, the genetic information of chADAMTS8 gene, its expression in chicken tissues, and chicken DF-1 cells under the stimulation of TLR3 were shown. The result suggests that chADAMTS8 expression may be induced by viral infection and correlated with TLR3-mediated signaling pathway. Further study of the function of chADAMTS8 during TLR3-dependent inflammation (which represents RNA viral infection) is needed and it will also be important to examine the molecular mechanisms during different regulation, depending on innate immune receptor activation.
A disintegrin and metalloproteinase with thrombospondin motifs (ADAMTSs) are secreted, extracellular enzymes with a compound domain organization [1]. In humans, nineteen ADAMTS proteins are encoded in the genome and they can be clustered into eight ŌĆścladesŌĆÖ based on their domain organization and known functions. The aggrecanase and proteoglycanase clades (ADAMTS1, 4, 5, 8, 9, 15, and 20) can cleave hyaluronan-binding chondroitin sulfate proteoglycans, a type of extracellular proteins, including aggrecan, versican, brevican, and neurocan [2]. Among them, ADAMTS8, also known as metalloprotease and thrombospondin domains 2 (METH-2), is a member of the ADAMTS family and was originally identified as an antiangiogenic factors that is down-regulated in brain cancer [3,4]. ADAMTS8 down-regulation has been also reported in other cancers, including brain, breast, lung, pancreatic, and thyroid cancer [4ŌĆō7]. The ADAMTS8 gene shows a high frequency of promoter methylation in brain, lung, and thyroid cancer, suggesting that the epigenetic silencing of ADAMTS8 may be involved in tumorigenesis [4,6,8]. Moreover, in cancer cells regulated by ADAMTS8, certain mutations can drive abnormal signaling during viral infection, indicating a correlation between cancers and viruses [9,10].
Pathogen-associated molecular patterns (PAMPs), such as polyinosinic-polycytidylic acid (poly[I:C]) and lipopolysaccharide, have been used to study toll-like receptor (TLR)-mediated cellular responses. Poly(I:C) is a synthetic analogue of double-stranded RNA (dsRNA), a PAMP generated during the replication of RNA and DNA viruses [11], and is recognized by distinct receptors depending on their localization. When added to the culture medium, poly(I:C) is mainly sensed by endosome-localized TLR3 [12,13]. TLR3 signaling can also occur in non-immune cells, contributing to an antitumor response. TLR3 is activated by extracellular dsRNA, which is recognized by the receptor in a sequence-independent manner. TLR3 initiates a protective response against dsRNA viruses including polio virus, coxsackievirus group B and serotype 3, and encephalomyocarditis virus, as well as DNA viruses, such as herpes simplex virus 1 and murine cytomegalovirus [14ŌĆō16]. Besides the mammals, TLR3 recognizes dsRNA virus infection and is involved in the resistance or susceptibility to viral infection in fowls. It was reported that the mRNA-expression levels of duck TLR3 and other cytokines (including interferon-╬▒ [IFN-╬▒]) were highly up-regulated during infection by duck reovirus, a dsRNA virus [17]. In 2014, Cheng et al [18] reported that infection with the chicken Newcastle disease virus increased the mRNA expression of chicken TLR3. ChTLR3 actively participates in the recognition of pro-inflammatory responses during viral infection, and leads to consequent antiviral cytokine secretion in chickens. In addition, TLR3 recognizes dsRNA that has been transcribed in vitro and its synthetic analogues, such as poly(I:C) and polyadenylic:polyuridylic acid (poly [A:U]). These analogues have been used to mimic responses to RNA virus infection and are commonly administered in in vitro and in vivo studies of TLR3-mediated cellular responses [19].
Although the expression of chicken ADAMTS8 (chADAMTS8) appears to be related to virus infection, the structure and expression pattern of this gene have not been studied. Here, we analyzed the amino acid sequence encoded by the chADAMTS8 gene by comparing it with the ADAMTS8 amino acid sequences from other species. Gene expression was investigated in various chicken tissues. In addition, the expression pattern of the chADAMTS8 gene was evaluated in the chicken DF-1 fibroblast cell line after stimulation with poly(I:C).
The chicken DF-1 cell line was purchased from the American Tissue Culture Collection (CRL-12203, Manassas, VA, USA). DF-1 cells were cultured in DulbeccoŌĆÖs modified EagleŌĆÖs medium supplemented with 10% fetal bovine serum, 2 mM L-glutamine, and 100 U/mL each of penicillin and streptomycin (Thermo Scientific, Logan, UT, USA) at 37┬░C in a humidified atmosphere of 5% CO2 and 95% air. Poly(I:C) was purchased from Invivogen (San Diego, CA, USA) and used for stimulation at a concentration of 10 ╬╝g/mL for 24 h.
The ADAMTS8 mRNA and amino acid sequences of various species (chicken, duck, human, chimpanzee, mouse, rat, cow, pig, and horse) were retrieved from the Ensembl database (http://www.ensembl.org/) (Table 1) and aligned with the BioEdit software, using the ClustalW method. The protein domains were predicted by using the SMART domain search program (http://smart.embl-heidelberg.de/). Phylogenetic analyses were performed with the MEGA7 software [20].
Trizol (Invitrogen, Carlsbad, CA, USA) was used to extract total RNA from tissue samples (liver, lung, kidney, spleen, and heart) and DF-1 cells. Total RNA was quantified using a NanoDrop spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). The chADAMTS8, chTLR3, and chTLR4 transcripts were analyzed by reverse transcription-polymerase chain reaction (RT-PCR) amplification. The RT-PCR conditions were as follows: an initial step of 94┬░C for 10 min; 35 cycles of 94┬░C for 30 s, 60┬░C for 30 s, and 72┬░C for 30 s; and a final step of 72┬░C for 10 min. The RT-PCR products were analyzed by electrophoresis on a 2.0% SeaKem LE agarose gel (Lonza, Basel, Switzerland). Target gene expression was normalized against that of the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene. The sequences of the GAPDH primers were 5ŌĆ▓-TGC TGC CCA GAA CAT CAT CC-3ŌĆ▓ for the forward primer and 5ŌĆ▓-ACG GCA GGT CAG GTC AAC AA-3ŌĆ▓ for the reverse primer.
Quantitative RT-PCR (qRT-PCR) was conducted with a CFX-96 RT-PCR detection system (BioRad, Hercules, CA, USA) to analyze chADAMTS8, chTLR3, chTLR4, and chIL1B expression. The sequences of the chADAMTS8 primers were 5ŌĆ▓-GCA CTA TGA CAC TGC CAT CCT-3ŌĆ▓ for the forward primer and 5ŌĆ▓-CGT GTC GCA GCC TTG ATG-3ŌĆ▓ for the reverse primer. ChTLR3 primers were: 5ŌĆ▓-CCA TTT TGA AGG GTG GAG AA-3ŌĆ▓ (forward) and 5ŌĆ▓-CCT GCT TCG AAG TCT CGT TC-3ŌĆ▓ (reverse). chTLR4 primers were: 5ŌĆ▓-TTC CAA GCA CCA GAT AGC AAC ATC-3ŌĆ▓ (forward) and 5ŌĆ▓-ACG GGT CAC AGA AGA ACT TAG GG-3ŌĆ▓ (reverse). chIL1B primers were: 5ŌĆ▓-GGA TTC TGA GCA CAC CAC AGT-3ŌĆ▓ (forward) and 5ŌĆ▓-TCT GGT TGA TGT CGA AGA TGT C-3ŌĆ▓ (reverse). The PCR conditions were as follows: an initial step of 94┬░C for 3 min; 39 cycles of 94┬░C for 10 s, 60┬░C for 30 s, and 72┬░C for 30 s; and a final step of 72┬░C for 10 min. Dissociation was performed at 0.5┬░C increments from 55┬░C to 95┬░C over 25 min. All samples were measured in triplicate to ensure reproducibility, and Ct values were calculated by the 2ŌłÆ╬ö╬öCt method [21]. Expression of the GAPDH gene was detected as the reference.
Results are presented as the means┬▒standard deviation of triplicate independent experiments. Statistical significance was assessed using a StudentŌĆÖs t-test. A p value of <0.05, compared with the non-treated control, was considered to reflect a statistically significant difference.
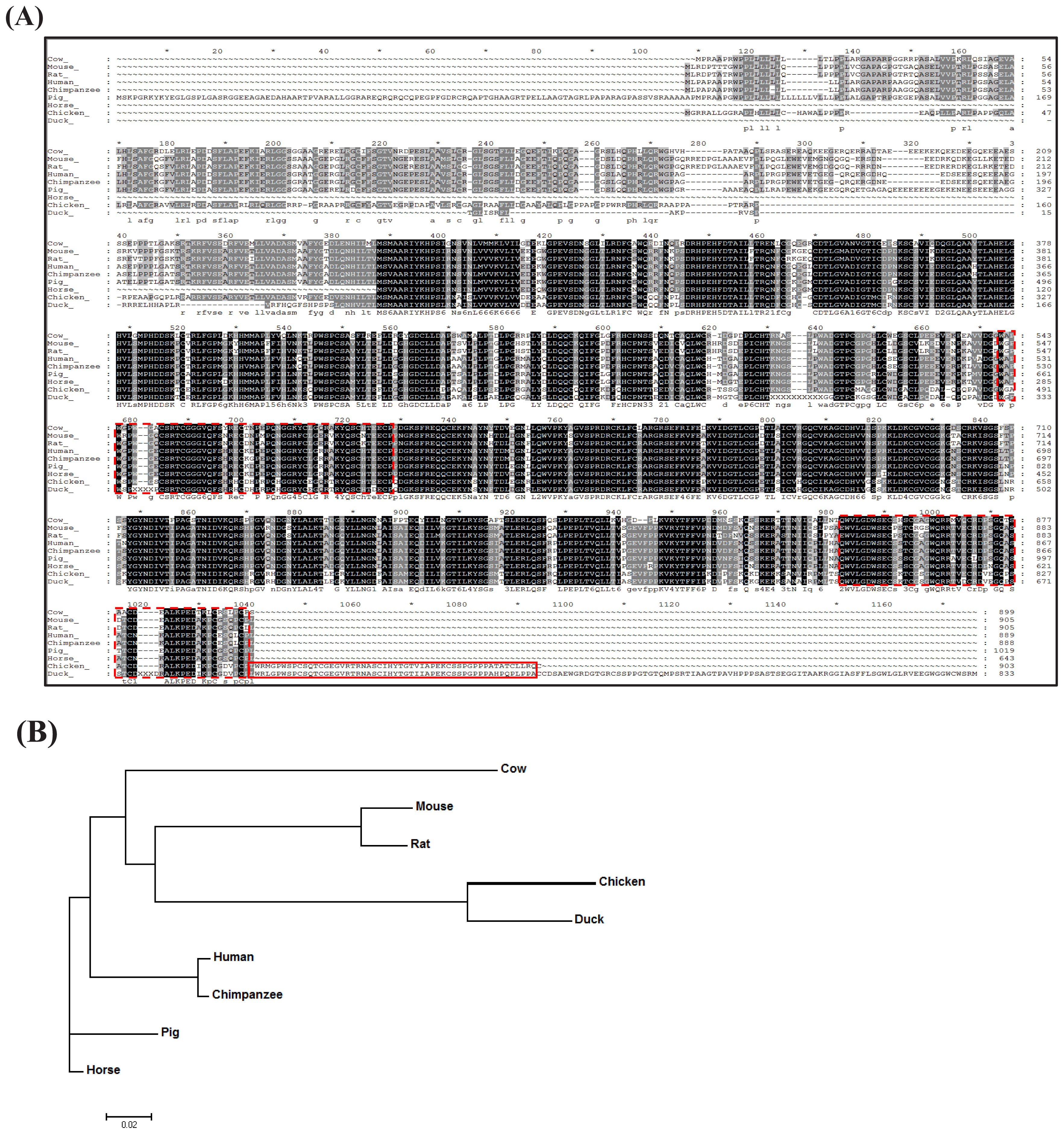
The amino acid sequence encoded by chADAMTS8 was analyzed and compared with that of the other species. ChADAMTS8 was identified as a differentially expressed gene (DEG) from chicken kidney by RNA-Seq analysis, after the chickens were fed a diet containing different amounts of calcium [22]. The chADAMTS8 amino acid sequence was compared with the duck, horse, pig, cow, mouse, rat, chimpanzee, and human sequences, which were retrieved from the Ensembl database. ChADAMTS8 was predicted to contain three thrombospondin type 1 (TSP1) domains, whose amino acid sequences were conserved among the species examined, whereas sequences outside of these domains (especially in the amino-terminal region) were diverse (Figure 1A). Specifically, all three TSP1 domains of duck ADAMTS8 shared homology with chADAMTS8, whereas only two TSP1 domains were homologous with those of the other mammal species. Nonetheless, this result suggests that the chADAMTS8 may serve similar biological functions as ADAMTS8 in other species, including antiangiogenic activity and tumorigenesis. The phylogenetic tree of ADAMTS8 revealed that chADAMTS8 is evolutionarily clustered in the same clade with that of the duck (Figure 1B).
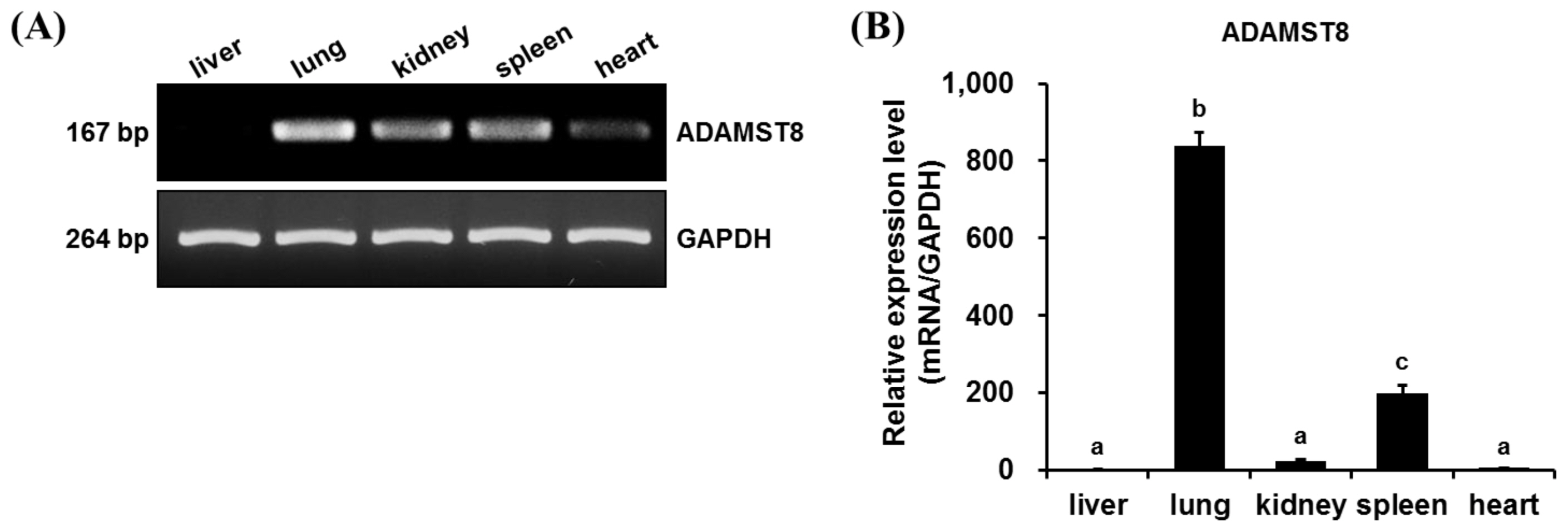
ADAMTS8 is known to be related to the development of lung cancer and recently, Zhou et al [23] reported that ADAMTS8 can be used as a biomarker for human lung cancer [6]. The gene-expression pattern of chADAMTS8 was investigated in various chicken tissues. RT-PCR and qRT-PCR analysis revealed that chADAMTS8 gene was highly expressed in the chicken lung tissue under normal conditions (Figure 2A, 2B). In addition to lung tissue, chADAMTS8 was also expressed in other tissues including spleen, kidney, heart, and liver. Like human ADAMTS8 gene expression in normal adult and fetal lung tissues, chADAMTS8 showed the highest expression in lung tissue and the second-highest expression in the spleen tissue. So far, it is not clear what the physiological role of chADAMTS8 is even though its expression is the highest in lung, and further study warrants revealing its role in normal condition as well as inflammatory conditions, caused by viral pathogens.
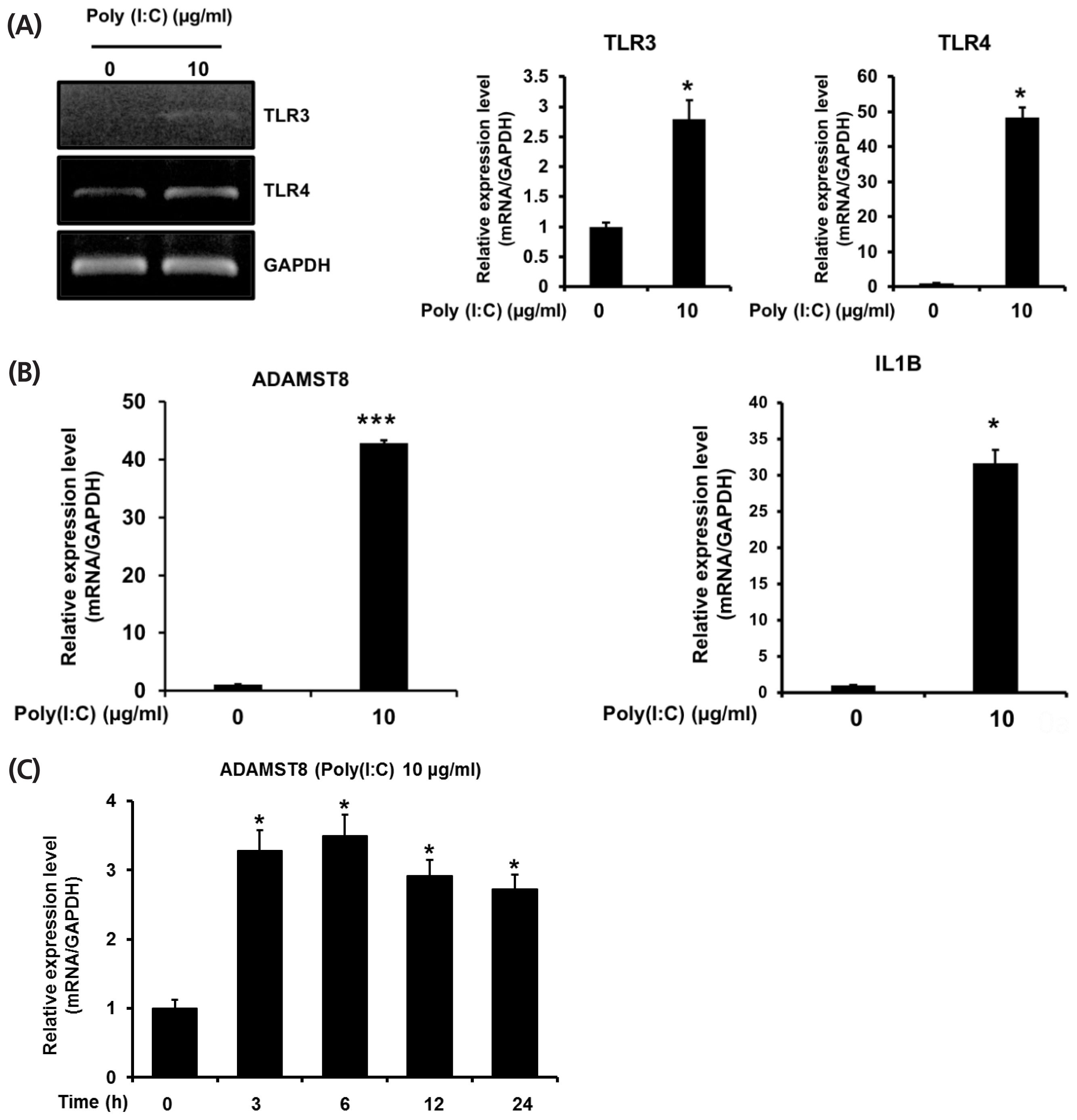
In addition, to gain insight into the regulation of chADAMTS8 gene expression, we examined its expression in chicken DF1 cells that were stimulated with poly(I:C). To confirm the innate immune responses of chicken DF-1 cell is stimulated by poly(I:C), the activation of TLR3 and TLR4 was examined with the agonist treatment. As shown in Figure 3A, the expressional levels of TLR3 and TLR4 in chicken DF-1 cell were increased under stimulation of poly(I:C). Additionally, the downstream activation of TLR3 pathway in response to the poly(I:C) stimulation was observed with interleukin 1 beta expression which is a cytokine induced following TLR3 activation (Figure 3B). A recent study reported that DF-1 cells express higher level of suppressor of cytokine signaling 1 (chSOCS1), a negative regulator of cytokine signaling in mammals, than chicken embryonic fibroblast cells, suggesting the dampened signaling activity of DF-1 cells in IFN-alpha (IFN-╬▒) signaling pathways through SOCS1 by inhibiting Janus kinase (JAK)-signal transducers and activator of transcription (STAT) signaling axes [24]. Nonetheless, our results show that chTLR3, chTLR4, and chIL1b expressions, as TLR3 signal target genes, are induced in poly(I:C) stimulation, suggesting that TLR3 signaling pathway of DF-1 cells may be separate from JAK-STAT signaling pathway activated by IFN-╬▒,╬▓, although further study is required to address it. Figure 3C shows qRT-PCR results of ADAMTS8 expression in chicken DF-1 cells with the treatment of poly(I:C). After TLR3/Mda5 agonist poly(I:C) stimulation, ADAMTS8 expression was measured time-dependently at 3, 6, 12, and 24 h. The expressional level of chADAMTS8 was increased as early as 3 h, and after that, the level was not elevated further until 24 h. Overall, these results suggest that chADAMTS8 expression may respond specifically to viral infection and thus, be correlated with TLR3-mediated cellular responses.
The complete molecular characterization of ADAMTS8 will be challenging; however, the genetic information and expression patterns of chADAMTS8 presented here can provide the foundation for further study including the transcriptional inhibition effects on the chADAMTS8 expression under the agonists poly(I:C). In addition, the up-regulation of chADAMTS8 after poly(I:C) treatment suggests that this gene might be related to viral infection in chickens and that chADAMTS8 gene is a promising candidate biomarker for infectious diseases that are rampant in chickens.
ACKNOWLEDGMENTS
This work was supported by the Next-Generation BioGreen 21 Program (No. PJ01324201, PJ01315101), Rural Development Administration, Republic of Korea and the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET) through the Agriculture, Food and Rural Affairs Research Center Support Program, funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA; grant number 716002-7).
Figure┬Ā1
Analysis of a disintegrin and metallopeptidase with thrombospondin motifs type 8 (ADAMTS8) amino acid sequences. (A) Comparison of the chicken ADAMTS8 (chADAMTS8) amino acid sequence with those of various other species. ChADAMTS8 was predicted to comprise three thrombospondin type-1 (TSP1) domains. Two domains (dashed box) were conserved well among the species, while the additional third domain (solid box) was homologous only with the duck sequence. (B) Phylogenetic tree for ADAMTS8 from various species. Phylogenetic analyses were performed with the amino acid sequence of each species, using MEGA7 software. ChADAMTS8 clustered in the same clade with duck. The bar indicates 2% amino acid divergence.
Figure┬Ā2
Expression of A disintegrin and metallopeptidase with thrombospondin motifs type 8 (ADAMTS8) in various chicken tissues. (A) Reverse-transcription polymerase chain reaction (RT-PCR) analysis of chADAMTS8 gene expression in various chicken tissues (liver, lung, kidney, spleen, and heart). (B) Quantitative PCR (qPCR) analysis of chADAMTS8 expression in chicken tissues. Relative expression levels were calculated using the 2ŌłÆ╬ö╬öCt method. The values are presented as the mean┬▒standard error of the mean. The expression levels of the gene varied among different tissues (n = 3, p<0.05). Significant differences were determined by TukeyŌĆÖs test, and bars with the same letter indicate cases where significant differences were not found (alpha = 0.05).
Figure┬Ā3
(A) Analyses of chTLR3, chTLR4, chIL1B, and chADAMTS8 expression in polyinosinic-polycytidylic acid (poly[I:C]) treated DF-1 cells by quantitative polymerase chain reaction (qPCR). Expression of chTLR3 and chTLR4, (B) chADAMTS8 and chIL1B in chicken DF-1 cells after poly(I:C) treatment. (C) DF-1 cells were treated with 10 ╬╝g/mL of poly(I:C) for 24 h (n = 3, * p<0.05). Time dependent expression of chADAMTS8 in poly(I:C) treated DF-1 cells. chTLR, chicken toll-like receptor; chIL1B, chicken interleukin 1 beta; chADAMTS8, chicken a disintegrin and metallopeptidase with thrombospondin motifs type 8.
Table┬Ā1
Ensembl and amino acid sequence IDs of ADAMTS8 genes of various species
REFERENCES
1. Kelwick R, Desanlis I, Wheeler GN, Edwards DR. The ADAMTS (A Disintegrin and Metalloproteinase with Thrombospondin motifs) family. Genome Biol 2015; 16:113



2. Stanton H, Melrose J, Little CB, Fosang AJ. Proteoglycan degradation by the ADAMTS family of proteinases. Biochim Biophys Acta 2011; 1812:1616ŌĆō29.


3. Vazquez F, Hastings G, Ortega MA, et al. METH-1, a human ortholog of ADAMTS-1, and METH-2 are members of a new family of proteins with angio-inhibitory activity. J Biol Chem 1999; 274:23349ŌĆō57.


4. Dunn JR, Reed JE, du Plessis DG, et al. Expression of ADAMTS-8, a secreted protease with antiangiogenic properties, is downregulated in brain tumours. Br J Cancer 2006; 94:1186ŌĆō93.




5. Porter S, Scott SD, Sassoon EM, et al. Dysregulated expression of adamalysin-thrombospondin genes in human breast carcinoma. Clin Cancer Res 2004; 10:2429ŌĆō40.


6. Dunn JR, Panutsopulos D, Shaw MW, et al. METH-2 silencing and promoter hypermethylation in NSCLC. Br J Cancer 2004; 91:1149ŌĆō54.




7. Masui T, Hosotani R, Tsuji S, et al. Expression of METH-1 and METH-2 in pancreatic cancer. Clin Cancer Res 2001; 7:3437ŌĆō43.


8. Rodriguez-Rodero S, Fernandez AF, Fernandez-Morera JL, et al. DNA methylation signatures identify biologically distinct thyroid cancer subtypes. J Clin Endocrinol Metab 2013; 98:2811ŌĆō21.



9. Kasuya H, Takeda S, Nomoto S, Nakao A. The potential of oncolytic virus therapy for pancreatic cancer. Cancer Gene Ther 2005; 12:725ŌĆō36.



10. Wennier S, Li S, McFadden G. Oncolytic virotherapy for pancreatic cancer. Expert Rev Mol Med 2011; 13:e18



11. Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol 2006; 80:5059ŌĆō64.



12. Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 2001; 413:732ŌĆō8.



13. Nishiya T, Kajita E, Miwa S, Defranco AL. TLR3 and TLR7 are targeted to the same intracellular compartments by distinct regulatory elements. J Biol Chem 2005; 280:37107ŌĆō17.


14. Oshiumi H, Okamoto M, Fujii K, et al. The TLR3/TICAM-1 pathway is mandatory for innate immune responses to poliovirus infection. J Immunol 2011; 187:5320ŌĆō7.


15. Abe Y, Fujii K, Nagata N, et al. The toll-like receptor 3-mediated antiviral response is important for protection against poliovirus infection in poliovirus receptor transgenic mice. J Virol 2012; 86:185ŌĆō94.



16. Negishi H, Osawa T, Ogami K, et al. A critical link between Toll-like receptor 3 and type II interferon signaling pathways in antiviral innate immunity. Proc Natl Acad Sci USA 2008; 105:20446ŌĆō51.



17. Zhang M, Song K, Li C, et al. Molecular cloning of Peking duck Toll-like receptor 3 (duTLR3) gene and its responses to reovirus infection. Virol J 2015; 12:207



18. Cheng J, Sun Y, Zhang X, et al. Toll-like receptor 3 inhibits Newcastle disease virus replication through activation of pro-inflammatory cytokines and the type-1 interferon pathway. Arch Virol 2014; 159:2937ŌĆō48.


19. Bianchi F, Pretto S, Tagliabue E, Balsari A, Sfondrini L. Exploiting poly(I:C) to induce cancer cell apoptosis. Cancer Biol Ther 2017; 18:747ŌĆō56.



20. Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 2016; 33:1870ŌĆō4.




21. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(ŌłÆDelta Delta C(T)) method. Methods 2001; 25:402ŌĆō8.


22. Park W, Rengaraj D, Kil DY, et al. RNA-seq analysis of the kidneys of broiler chickens fed diets containing different concentrations of calcium. Sci Rep 2017; 7:11740










 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print





