 |
 |
| Anim Biosci > Volume 31(8); 2018 > Article |
|
Abstract
Objective
This study was designed to investigate the effects of temperature and storage time on the evolution of bacterial communities in swine manure.
Methods
Manure was stored at ŌłÆ20┬░C, 4┬░C, 20┬░C, or 37┬░C and sampled at 7-day intervals over 28 days of storage, for a total of 5 time points. To assess the bacterial species present, 16S ribosomal RNA gene sequences were analyzed using pyrosequencing.
Results
After normalization, 113,934 sequence reads were obtained, with an average length of 466.6┬▒4.4 bp. The diversity indices of the communities reduced as temperature and storage time increased, and the slopes of rarefaction curves decreased from the second week in samples stored at ŌłÆ20┬░C and 4┬░C. These results indicate that the richness of the bacterial community in the manure reduced as temperature and storage time increased. Firmicutes were the dominant phylum in all samples examined, ranging from 89.3% to 98.8% of total reads, followed by Actinobacteria, which accounted for 0.6% to 7.9%. A change in community composition was observed in samples stored at 37┬░C during the first 7 days, indicating that temperature plays an important role in determining the microbiota of swine manure. Clostridium, Turicibacter, Streptococcus, and Lactobacillus within Firmicutes, and Corynebacterium within Actinobacteria were the most dominant genera in fresh manure and all stored samples.
Conclusion
Based on our findings, we propose Clostridium as an indicator genus of swine manure decomposition in an anaerobic environment. The proportions of dominant genera changed in samples stored at 20┬░C and 37┬░C during the fourth week. Based on these results, it was concluded that the microbial communities of swine manure change rapidly as storage time and temperature increase.
The various microorganisms that are present in swine slurry, such as Bacillus, Clostridium, and Lactobacillus, play a vital role in the decomposition of manure. These organisms live in a partially anaerobic environment and therefore utilize a variety of potential substrates, including proteins and non-starch polysaccharides [1]. The pH of swine manure is relatively stable because of low lactate concentration, and microbial activity is mainly affected by the buffering capacity of ammonia nitrogen [2]. Therefore, environmental conditions, such as temperature, moisture, and oxygen availability determine the rate and extent of microbial-mediated substrate conversion into various compounds [3].
Fecal bacteria are often used as an indicator of fecal contamination in aqueous and solid samples. Consequently, traditional biological methods used to detect fecal pollution have relied on bacterial indicators. Total coliforms, Escherichia coli (E coli), fecal streptococci, enterococci, Clostridium perfringens (C. perfringens), and bifidobacteria have all been suggested to be key fecal indicator microorganisms [4]. Of these, E. coli is considered the most accurate indicator of contamination by human and animal waste, as 90% to 100% of the isolated coliform microorganisms from human and animal feces are E. coli [5]. Detailed characterization of the intestinal microbiota of the swine intestine using anaerobic culturing techniques has revealed that the major bacterial groups present are Streptococcus, Lactobacillus, Prevotella, Selenomonas, Mitsuokella, Megasphaera, Clostridium, Eubacterium, Bacteroides, Fusobacterium, Acidaminococcus, and Enterobacteriaceae [6]. These studies highlight potential enteric viruses and indicator organisms commonly found in fecal samples from watershed animals that could serve as an indicator of fecal contamination.
Bacterial diversity and dynamics have been found to alter in swine manure during the acidification process [7,8]. These studies primarily identified microbes using polymerase chain reaction (PCR)-denaturing gradient gel electrophoresis (DGGE) [7]. The species most typically identified based DGGE analysis are Acinetobacter lwoffii, Alcaligenes spp., Bacteroides spp., Botryosphaeria dothidea, Comamonas spp., Pseudomonas spp., and Hydrogenophaga spp. Tracking pathogenic or fecal indicators in animal waste through the application of cell culture has also been reported [8]. Thorough analysis of microbial population dynamics using molecular methods may lead to a better understanding of the metabolic transformations that occur in manure [9]. Various molecular markers have been developed for the identification and quantification of eubacteria from the swine intestinal tract, including specific 16S rRNA gene sequences [10]. Gourmelon et al [11] identified and quantified the BacteroidesŌĆōPrevotella group in different preserved manure samples by using specific markers (PF163F/Bac708R for Bacteroidales). Using Lab 158 probe and Bac 303 probe, microaerophilic species, lactobacilli (e.g., L. amylovorus), and Bacteroides fragilis have been detected in piglet feces [12].
However, the diversity and evolution of microorganisms during the process of swine manure decomposition have not been well studied. To address this, the present study investigated the evolution of microorganism communities that associate with the decomposition of pig manure over different time periods and at various temperatures using pyrosequencing to identify the specific organisms responsible for each phase of decomposition.
Swine manure samples (approximately 10 kg) were collected from the scraper-type experimental porker farm of the National Institute of Animal Science, Korea. The collected manure was homogenized by passing it through a 2 mm├Ś2 mm sieve and was squeezed with gauze to remove remaining debris. Forty milliliters of the homogenate was transferred into a 50-mL Falcon tube (C2602, MTC Bio, Metuchen, NJ, USA). In total, 51 samples were prepared, including 48 samples that were divided into 4 temperature treatment groups (n = 12 each) and were stored at ŌłÆ20┬░C, 4┬░C, 20┬░C, or 37┬░C. Samples at ŌłÆ20┬░C and 4┬░C were stored in a combined freezer/refrigerator (SR6449M; Samsung, Suwon, Korea). Samples at 20┬░C and 37┬░C were stored in an incubator (VS-1203P; Vision Scientific, Daejeon, Korea). For 4 weeks, at the end of each week, 3 samples were collected for each condition. Three of the 51 samples were analyzed immediately (time point 0, referred to as ŌĆ£fresh manure samplesŌĆØ). Each sample was analyzed by pyrosequencing in triplicate.
Metagenomic DNA was extracted from treated swine manure samples using a Fast DNA spin kit for soil (MP Biomedicals, Santa Ana, CA, USA). The extracted DNA was purified using a PowerClean DNA Clean-up Kit (Mo Bio, Carlsbad, CA, USA), according to the manufacturerŌĆÖs instructions. A fragment of the bacterial 16S rRNA gene (V1ŌĆōV3 region) was amplified from each purified DNA sample using bacterial fusion primers [13]. Each amplification mixture (50 ╬╝L) consisted of 10├Ś Taq buffer, dNTP mixture (TaKaRa, Shiga, Japan), 10 ╬╝M each primer, 2 U of Taq polymerase (Ex Taq; TaKaRa, Japan) and 2 ╬╝L of the DNA template. The reactions were carried out in a C1000 Touch thermocycler (Bio-Rad, Hercules, CA, USA) [14]. Amplified products were confirmed by gel electrophoresis on a GelDoc system (Bio-Rad, USA) and then purified using a QIAquick PCR Purification Kit (Qiagen, Valencia, CA, USA). Purified amplicons from 17 distinct different samples stored at different temperatures and times (fresh sample and 16 conditions) were pooled at the same concentration. Fragments shorter than the target region were removed using an AMPure Bead Kit (Agencourt Bioscience, Beverly, MA, USA). Purified products were amplified on sequencing beads by emulsion PCR. Beads were recovered from emulsion PCR and DNA was sequenced using a GS Junior system (Roche, Branford, CT, USA), following the manufacturerŌĆÖs instructions. Pyrosequencing was conducted at Chunlab Inc. (Seoul, Korea).
Pyrosequencing data were analyzed using a previously described protocol [13,14]. Raw sequencing reads were sorted by their unique barcodes and primer sequences were trimmed using the HMMER 3.0 package [15]. Low-quality reads (average quality score <25 or read length <300 bp) were filtered and removed from further analysis. Sequencing errors were corrected by read clustering, and representative sequences were used for taxonomic identification. Each representative read was assigned a taxonomic position based on the highest pairwise similarity value using the EzTaxon-e database [16]. Chimeric sequences were removed using UCHIME [17]. Random subsampling was used to normalize read numbers in the samples as different read numbers were obtained for each sample, which can affect statistical calculations. Statistical estimations of the Chao1 index, Shannon diversity index, and GoodŌĆÖs coverage were performed at 3% dissimilarity, using mothur [18]. Relationships between samples were evaluated by principal coordinate analysis (PCoA) using the Fast UniFrac distance [19]. The significance of differences in bacterial communities among samples was calculated using Libshuff analysis, while the similarity coefficients of BrayŌĆōCurtis, Jaccard abundance, and Sorenson abundance were obtained using mothur. The pyrosequencing read data obtained in this study are deposited at the European Molecular Biology Laboratory (EMBL).
The present study assessed, for the first time, the effects of incubation time and temperature on the evolution of bacterial communities in swine manure using 16S rRNA gene pyrosequencing. In total, data from 51 distinct samples, including fresh manure, were analyzed and compared. The estimated diversity indices obtained after normalization are shown in Table 1. In total, 113,934 reads, with an average length of 466.6┬▒4.4 bp, were obtained. The minimum and maximum coverage was 76% and 84%, respectively. The number of operational taxonomic units (OTUs) was greater than 1,000 in the original manure, although a similarly high number of OTUs was obtained only for the ŌłÆ20┬░C and 4┬░C samples during the first week of storage.
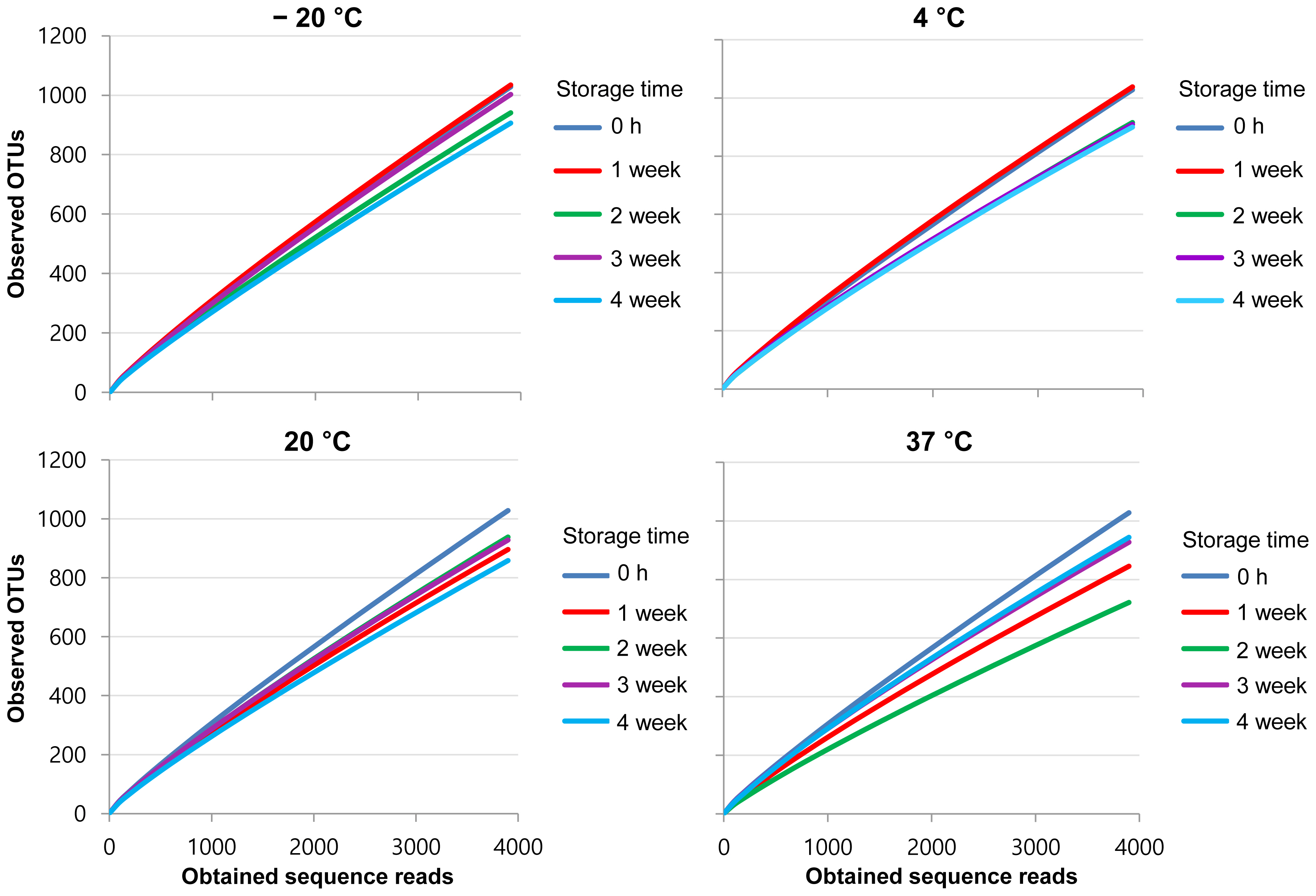
The diversity indices decreased with increasing temperature. Samples stored for 4 weeks at 20┬░C, and for all times when stored at 37┬░C, had lower diversity indices than the original sample. The lowest diversity index (3.36) was obtained for samples stored at 37┬░C for 2 weeks. To establish the number of species as a function of sample number, we plotted rarefaction curves, which showed a trend similar to that of the diversity indices (Figure 1). The rarefaction curves also revealed that the number of species initially increased rapidly as the most common species were found, but then plateaued as only the rarest species remained to be sampled [20]. The slopes of the curves decreased from the second week for samples stored at ŌłÆ20┬░C and 4┬░C, and the differences became more pronounced as the incubation time increased. The slopes significantly changed in the samples incubated at 20┬░C and 37┬░C. These results indicated that the richness of the bacterial community in the manure was reduced as storage temperature and time increased.
The compositions of the bacterial communities in each sample group were compared at the phylum and genus levels using double circle charts (Figure 2). Firmicutes was the dominant phylum in all samples (ranging from 89.3% to 98.8% of total reads), while Actinobacteria was the second dominant group in most of the samples (0.6% to 7.9%). The proportion of Proteobacteria increased in the samples stored for 4 weeks at 37┬░C (to 6.4%). However, the phylum-level composition of samples incubated at ŌłÆ20┬░C was similar to that in the original swine manure. For the samples stored at 4┬░C, the relative abundance of Actinobacteria increased after 3 weeks, while that of Firmicutes decreased. This indicated that the composition of a bacterial community can change even at relatively low temperatures. The composition of the community began to alter starting from the first week when incubated at 37┬░C, suggesting that temperature is an important factor that affects the microbiota in swine manure. The genera Clostridium, Turicibacter, Streptococcus, and Lactobacillus within Firmicutes, as well as Corynebacterium within Actinobacteria were dominant in the fresh manure and in most of the stored samples. The proportions of dominant genera changed in the samples stored for 4 weeks at 20┬░C and in all samples stored at 37┬░C. The proportion of Clostridium in the samples stored for 4 weeks at 20┬░C was higher than that in the other samples, while the ratio of Streptococcus to Lactobacillus decreased. Similar changes in genus composition were observed after 1 week in samples incubated at 37┬░C. The proportions of Streptococcus and Lactobacillus were reduced to 0.1% and 0.2% within the first two weeks at 37┬░C (12.2% and 11.0% in the fresh manure sample, respectively), while the proportion of Turicibacter increased by 19.7% and 23.7% after 1 and 2 weeks of storage, respectively. However, the abundance of Turicibacter decreased (to 14.6%) in the samples stored at 37┬░C from the third week, whereas uncultured bacteria, such as Bacillus and Turicibacter, increased after 3 to 4 weeks of storage. The relative abundance of Pusillimonas within Proteobacteria increased (3.5% of the total reads) after 4 weeks of storage at 37┬░C.
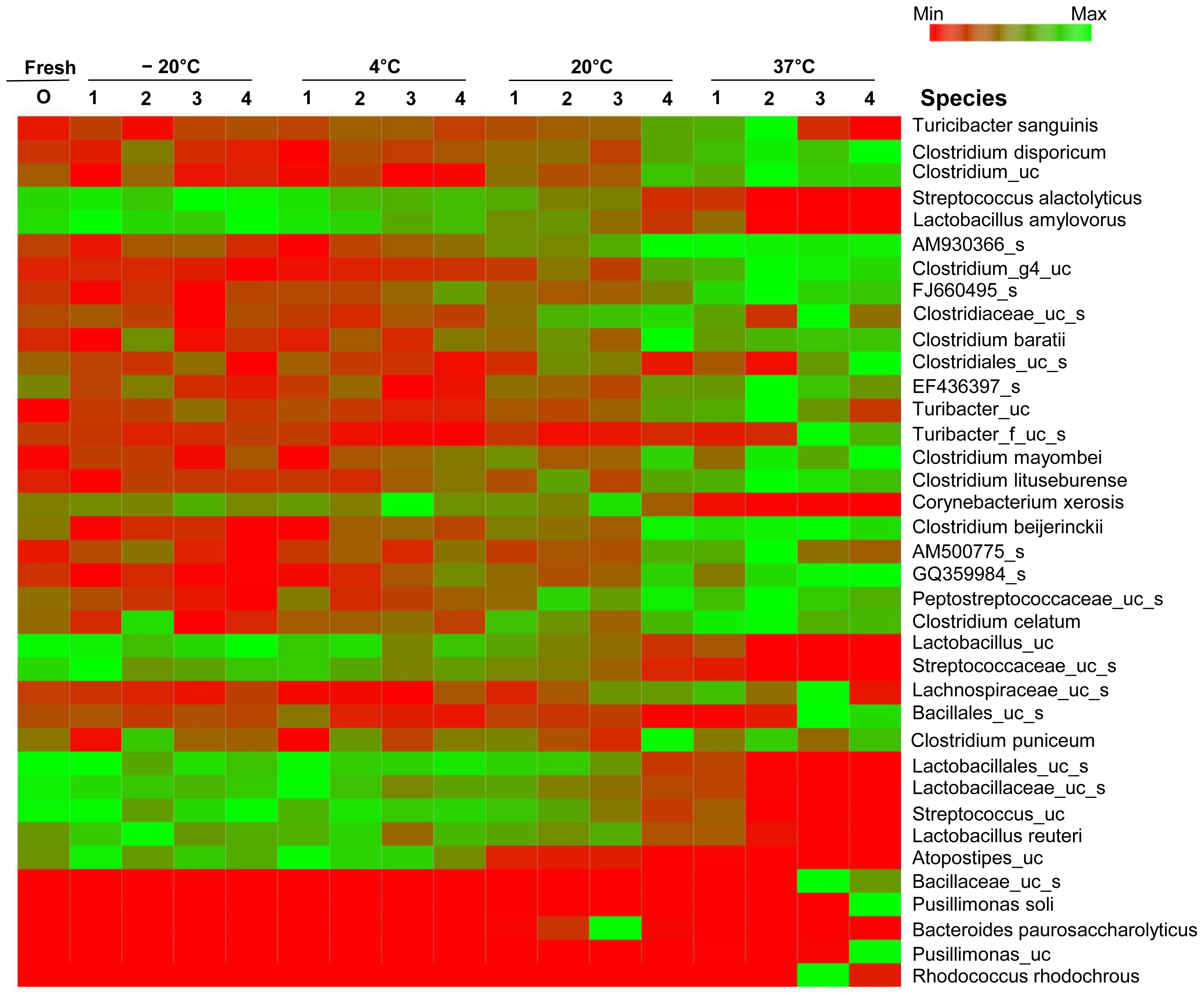
Detailed information on the influenced bacteria was obtained at the species level by heatmap analysis (Figure 3). The swine manure stored at the conditions of different temperature and time were dominated by order Clostridiales and family Clostridiaceae. The profile of fresh manure was similar to those of samples stored at ŌłÆ20┬░C and 4┬░C. The relative abundances of each species changed in samples stored at 20┬░C, and differences in the bacterial communities were observed in samples incubated at 20┬░C for 4 weeks, and in all samples incubated at 37┬░C. The relative abundances of Streptococcus alactolyticus, L. amylovorus, Corynebacterium xerosis, unclassified Lactobacillus, unclassified Streptococcus, Lactobacillus reuteri, and unclassified Atopostipes decreased as the incubation temperature and time increased. Streptococcus, Lactobacillus played an important role at the beginning of degradation of swine manure at low temperature. The proportions of T. sanguinis, Clostridium disporicum, Clostridium baratii, Terrisporobacter mayombei, Clostridium lituseburense, Clostridium beijerinckii, Clostridium celatum, Clostridium puniceum, and various uncultured Clostridium, Turibacter, and Bacillus increased in the samples incubated for 4 weeks at 20┬░C and in all samples incubated at 37┬░C. The bacterial communities of samples incubated at 37┬░C for 1 and 2 weeks were different from those in the samples incubated for 3 and 4 weeks. The relative abundances of T. sanguinis and unclassified bacteria similar to unclassified rumen bacterial clones EF436397 and AM500775 decreased from the third week of incubation at 37┬░C, whereas unclassified Turicibacter and Bacillus increased. Pusillimonas soli and unclassified Pusillimonas increased in the samples that were incubated for 4 weeks. Of these particular species, Clostridium disporicum has been associated with the production of skatole (3-methylindole) [20], and most Clostridium spp. produce strong odors.
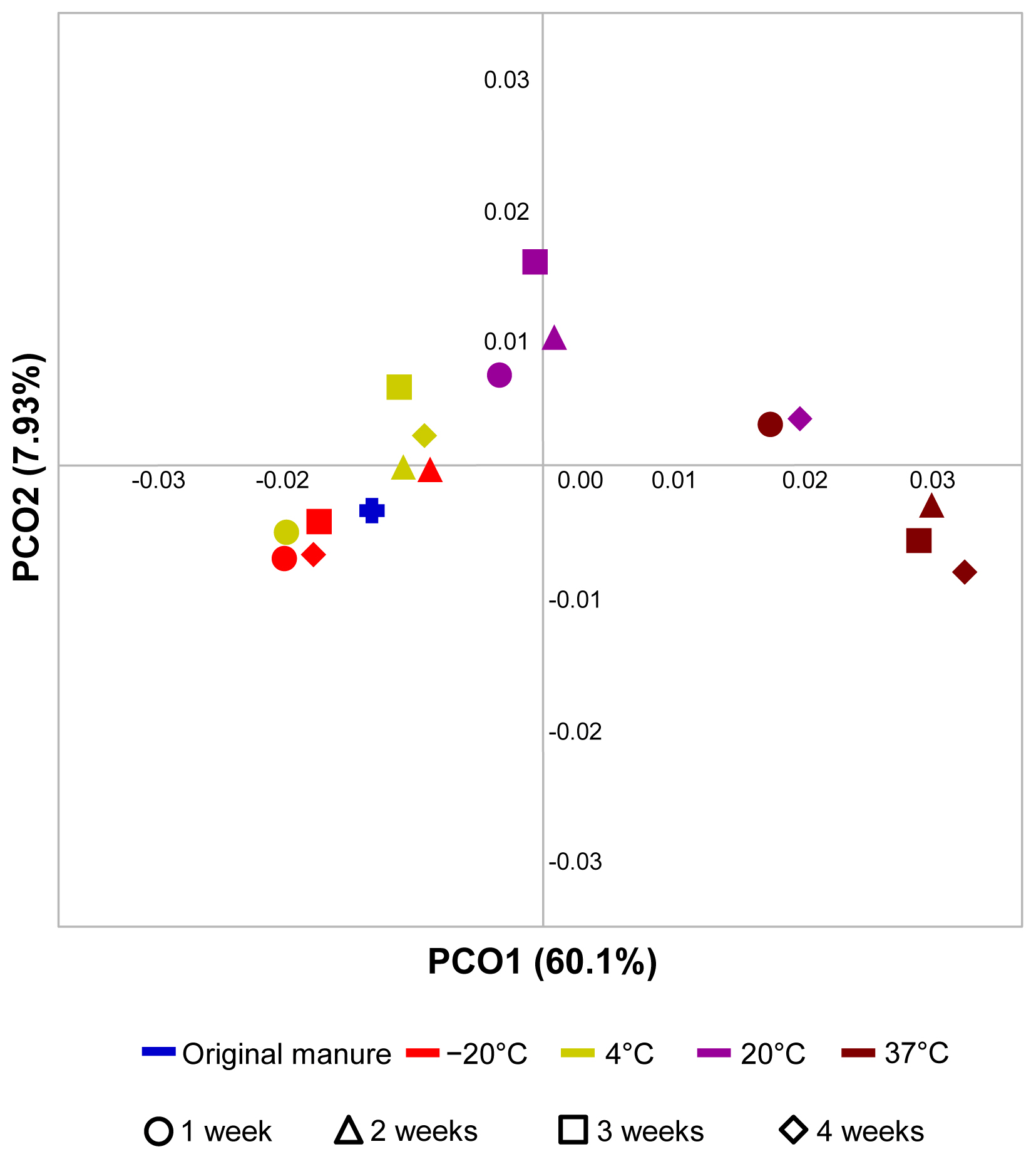
The differences in and relations among bacterial communities in swine manure under each treatment are shown in PCoA plots generated using Fast UniFrac distances (Figure 4). The bacterial communities of samples incubated at 20┬░C and 37┬░C were significantly different from the fresh manure. However, the UniFrac distances between fresh manure and samples incubated at ŌłÆ20┬░C and 4┬░C were smaller than those between the fresh manure and samples incubated at 20┬░C and 37┬░C. The difference between bacterial communities in the samples stored at 20┬░C as compared to fresh manure was the largest at 4 weeks. The UniFrac distances between bacterial communities in the samples incubated at 37┬░C were also greater than those between samples incubated at other temperatures. These distances increased with increased storage time at 37┬░C. In summary, this part of the study revealed that the microbial communities during manure decomposition changed rapidly as storage time and temperature increased, especially for manure that featured a predominance of Firmicutes and Actinobacteria.
In this study, Firmicutes were the dominant phylum in all samples examined, ranging from 89.3% to 98.8% of total reads, followed by Actinobacteria, which accounted for 0.6% to 7.9%. This finding is similar to those of Haakensen et al [21], who reported that Firmicutes are typically associated with animal manure and occur as contaminants that arise during food and brewing processes. Clostridium, Turicibacter, Streptococcus, and Lactobacillus within Firmicutes, and Corynebacterium within Actinobacteria were the most dominant genera in the fresh manure and stored samples. Clostridium [22], Streptococcus [23,24], and Lactobacillus [25,26] within Firmicutes have been considered indicators of animal fecal pollution in previous studies [27]. Turicibacter and Corynebacterium are newly suggested indicators. Clostridium is considered an indicator genus of swine manure decomposition and causes of manure odor in an anaerobic environment. Previous studies have indicated that swine fecal microbial communities appear to be anaerobic as the swine digestive tract is dominated by fermentative bacteria, such as low-G+C-content gram-positive bacteria belonging to the EubacteriumŌĆōClostridium and LactobacillusŌĆōStreptococcus groups, and gram-negative Bacteroides [27]. C. perfringens and its spores have been considered an important indicator of fecal pollution. Although the spores of C. perfringens are relatively easy to detect, they are also particularly robust and more tolerant to environmental effects than other traditional enteric indicators, making them ideal indicators of fecal contamination [22]. Streptococcus bovis is considered an indicator of bovine fecal pollution [23], but it is also commonly found in non-bovine ruminants and feces from other animals, including dogs, cats, horses, pigs, and various birds. The World Health Organization [24] recommends the use of fecal streptococci as an indicator of fecal pollution (WHO [24]). Additionally, Enterococcus spp. occur at high densities in animal feces. The enterococci are a sub-group of the fecal streptococcus group and consist of a number of species, including E. faecalis, E. faecium, S. gallinarum, and S. avium. Because of the high prevalence and abundance of L. amylovorus in piglet feces, researchers have proposed that this species may serve as a novel marker for pig fecal contamination [25]. Finally, Marti et al [26] examined 163 samples, including 48 pig and 87 other animal fecal samples, in addition to 28 human fecal samples, demonstrating that L. amylovorus was present in 100% of the swine fecal samples.
Kim [28] analyzed pH, NH4-N (%), and volatile fatty acids in samples of swine slurry collected from a farm over a course of 7 weeks. The analyses revealed changes from the initial concentrations of fatty acids in the first 4 weeks (acetic acid [880ŌåÆ2479 ppm], propionic acid [290ŌåÆ550 ppm], butyric acid [216ŌåÆ235 ppm], valeric acid [30ŌåÆ68 ppm], iso-butyric acid [157ŌåÆ249 ppm], and iso-valeric acid [210ŌåÆ349 ppm]). The highest concentrations (5,673, 1,155, 1,075, 94, 469, 540 ppm, respectively) were observed in the 6th week. The pH did not change significantly, from 6.86 to 7.29, during the period. In addition, NH4-N (%) did not significantly change from the initial concentration of 0.33. In other words, there were no significant differences in the physicochemical properties (pH or NH4-N [%]) of the swine slurry throughout storage.
In the current study, a decrease in bacterial community diversity was found to occur early, within the first 7 days, for samples stored at 37┬░C. However, the diversity loss occurred at a slower rate (after 4 weeks) in samples stored at 20┬░C. Based on the results obtained in the present study, it may be concluded that temperature plays an important role in the evolution of microbial communities in swine manure. Through controlling the temperature and storage period of manure and subsequently affecting the microbial communities, it may be possible to better control the odors typically generated by manure.
ACKNOWLEDGMENTS
This work was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET) through the Advanced Production Technology Development Program, funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA) (316012-03-2-WT011).
Figure┬Ā1
Rarefaction curves of the pyrosequenced bacterial communities present in swine manure. Rarefaction curves of the bacterial communities present in swine manure were compared across various storage temperatures (ŌłÆ20┬░C, 4┬░C, 20┬░C, and 37┬░C) and periods (0 h, 1 week, 2 weeks, 3 weeks, and 4 weeks) after normalizing the read numbers in each sample. Blue line represent the time point 0, red line represent 1st week, green line represent 2nd weeks, violet line represent 3rd weeks, and sky-blue line represent 4th weeks.
Figure┬Ā2
Bacterial community compositions. Bacterial community composition in each sample at the phylum and genus levels is shown using double circle charts. Inner circle represent phylum composition; outer circle represent genus composition.

Figure┬Ā3
Heatmap analysis. Heatmap analysis of each sample according to incubation time and temperature. The color code indicates the differences in the relative abundance from the mean, ranging from red (negative) through black (the mean) to the green (positive). uc, unclassified.
Figure┬Ā4
UniFrac distance-based clustering of bacterial communities associated with different storage time and temperature. Principal coordinates (PCoA) plots using weighted UniFrac distances for samples incubated under different conditions. The following UniFrac analysis were based on the operational taxonomic unit (OTU) data, with the first three principal coordinates. The fresh original manure sample as the control was included in this analysis. Blue line represent the original fresh manure, red line represent the manure stored in ŌłÆ20┬░C, green line represent the manure stored in 4┬░C, violet line represent the manure stored in 20┬░C, and plum line represent the manure stored in 37┬░C. Circle represent 1st week, triangle represent 2nd weeks, square represent 3rd weeks, and rhombus represent 4th weeks.
Table┬Ā1
Summary of estimated diversity indices after normalization
REFERENCES
1. Mackie RI, Stroot PG, Varel VH. Biochemical identification and biological origin of key odor components in livestock waste. J Anim Sci 1998; 76:1331ŌĆō42.


2. Switzenbaum MS, Giraldo-Gomez E, Hickey RF. Monitoring of the anaerobic methane fermentation process. Enzyme Microb Technol 1990; 12:722ŌĆō30.

3. Miller DN, Varel VH. Swine manure composition affects the biochemical origins, composition, and accumulation of odorous compounds. J Anim Sci 2003; 81:2131ŌĆō8.



4. Clark RM, Geldreich EE, Fox KR, et al. Tracking a Salmonella serovar typhimurium outbreak in Gideon, Missouri: role of contaminant propagation modelling. J Water Supply Res Technol 1996; 45:171ŌĆō83.
5. Hurst CJ, Crawford RL, Knudsen GR, et al. Manual of environmental microbiology. second edWashington, DC, USA: ASM Press; 2002.
6. Jensen BB. Possible ways of modifying type and amount of products from microbial fermentation in the gutPiva A, Bach Knudsen KE, Lindberg J-E, editorsGut environment of pigs. Loughborough, UK: Nottingham University Press; 2001. p. 181ŌĆō200.
7. Zhang D, Yuan X, Guo P, et al. Microbial population dynamics and changes in main nutrients during the acidification process of pig manures. J Environ Sci (China) 2011; 23:497ŌĆō505.


8. Munch B, Errebo Larsen H, Aalb├”ck B. Experimental studies on the survival of pathogenic and indicator bacteria in aerated and non-aerated cattle and pig slurry. Biol Wastes 1987; 22:49ŌĆō65.

9. Peu P, Brug├©re H, Pourcher A-M, et al. Dynamics of a pig slurry microbial community during anaerobic storage and management. Appl Environ Microbiol 2006; 72:3578ŌĆō85.



10. Okabe S, Shimazu Y. Persistence of host-specific Bacteroides-Prevotella 16S rRNA genetic markers in environmental waters: effects of temperature and salinity. Appl Microbiol Biotechnol 2007; 76:935ŌĆō44.


11. Gourmelon M, Caprais MP, S├®gura R, et al. Evaluation of two library-independent microbial source tracking methods to identify sources of fecal contamination in French estuaries. Appl Environ Microbiol 2007; 73:4857ŌĆō66.



12. Pieper R, Janczyk P, Zeyner A, et al. Ecophysiology of the developing total bacterial and Lactobacillus communities in the terminal small intestine of weaning piglets. Microb Ecol 2008; 56:474ŌĆō83.


13. Hur M, Kim Y, Song HR, et al. Effect of genetically modified poplars on soil microbial communities during the phytoremediation of waste mine tailings. Appl Environ Microbiol 2011; 77:7611ŌĆō9.



14. Kim BS, Kim JN, Yoon SH, et al. Impact of enrofloxacin on the human intestinal microbiota revealed by comparative molecular analysis. Anaerobe 2012; 18:310ŌĆō20.


16. Kim OS, Cho YJ, Lee K, et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 2012; 62:716ŌĆō21.


17. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera dectection. Bioinformatics 2011; 27:2194ŌĆō200.




18. Schloss PD, Westcott SL, Ryabin T, et al. Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 2009; 75:7537ŌĆō41.



19. Hamady M, Lozupone C, Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J 2010; 4:17ŌĆō27.



20. Andrew FS. Rarefraction curvesKotz S, Campbell B, Balakrishnan N, Vidakovic B, editorsEncyclopedia of Statistical Science. New York, USA: John Wiley & Sons, New York, USA; 2006.
21. Haakensen M, Dobson CM, Deneer H, et al. Real-time PCR detection of bacteria belonging to the Firmicutes phylum. Int J Food Microbiol 2008; 125:236ŌĆō41.


22. Tallon P, Magajna B, Lofranco C, et al. Microbial indicators of fecal contamination in water: a current perspective. Water Air Soil Pollut 2005; 166:139ŌĆō66.

23. Kator H, Rhodes MW. Microbial and chemical indicatorsHackney C, Pierson M, editorsEnvironmental indicators and shellfish safety. New York, USA and London, UK: Chapman and Hall Publishers; 1994. p. 30ŌĆō91.
24. World Health Organization. Guidelines for drinking water qualitysecond ed.2:Health criteria and other supporting information. Geneva, Switzerland: World Health Organization; 1996.
25. Marti R, Dabert P, Pourcher A-M. Pig manure contamination marker selection based on the influence of biological treatment on the dominant fecal microbial groups. Appl Environ Microbiol 2009; 75:4967ŌĆō74.



26. Marti R, Dabert P, Ziebal C, et al. Evaluation of Lactobacillus sobrius/L. amylovorus as a new microbial marker of pig manure. Appl Environ Microbiol 2010; 76:1456ŌĆō61.


27. Peu P, Brug├©re H, Pourcher A-M, et al. Dynamics of a pig slurry microbial community during anaerobic storage and management. Appl Environ Microbiol 2006; 72:3578ŌĆō85.



28. Kim WJ. Improvement of livestock odor through livestock manure recycling project. In : Fall Conference of the Korean Society of Odor Research and Engineering 2017; 2017 Nov 23ŌĆō24; Lee Tae-seok Memorial Hall, Inje University. Gimhae, Korea: Korean Society of Odor Research and Engineering; 2017. p. 44ŌĆō56.






 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print





