 |
 |
| Anim Biosci > Volume 31(6); 2018 > Article |
|
Abstract
Objective
The purpose of this study was to investigate the genetic effects of six keratin (KRT) genes on the wool traits of 418 Chinese Merino (Xinjiang type) (CMXT) individuals.
Methods
To explore the effects and association of six KRT genes on sheep wool traits, The polymerase chain reaction-based single-strand conformation polymorphism (PCR-SSCP), DNA sequencing, and the gene pyramiding effect methods were used.
Results
We report 20 mutation sites (single-nucleotide polymorphisms) within the six KRT genes, in which twelve induced silent mutations; five induced missense mutations and resulted in Ile→Thr, Glu→Asp, Gly→Ala, Ala→Ser, Se→His; two were nonsense mutations and one was a same-sense mutation. Association analysis showed that two genotypes of the KRT31 gene were significantly associated with fiber diameter (p<0.05); three genotypes of the KRT36 gene were significantly associated with wool fineness score and fiber diameter (p<0.05), three genotypes of the KRT38 gene were significantly associated with the number of crimps (p< 0.05); and three genotypes of the KRT85 gene were significantly associated with wool crimps score, body size, and fiber diameter (p<0.05). Analysis of the gene pyramiding effect between the different genotypes of the gene loci KRT36, KRT38, and KRT85, each genotype in a gene locus was combined with all the genotypes of another two gene loci and formed the different three loci combinations, indicated a total of 26 types of possible combined genotypes in the analyzed population. Compared with the other combined genotypes, the combinations CC-GG-II, CC-HH-IJ, CC-HH-JJ, DD-HH-JJ, CC-GH-IJ, and CC-GH-JJ at gene loci KRT36, KRT38, and KRT85, respectively, had a greater effect on wool traits (p<0.05).
Conclusion
Our results indicate that the mutation loci of KRT31, KRT36, KRT38, and KRT85 genes, as well as the combinations at gene loci KRT36, KRT38, and KRT85 in CMXT have significant effects on wool traits, suggesting that these genes are important candidate genes for wool traits, which will contribute to sheep breeding and provide a molecular basis for improved wool quality in sheep.
Sheep farming is an important component of animal husbandry, and many researchers in developed and developing countries are now considering it as a career. The efficiency of wool processing is dependent on the consistency of wool fiber. Although selective breeding of sheep has partly reduced the variation in wool fiber, considerable variation both within and between fleeces still exists [1]; therefore, accelerating the efficiency of wool breeding is very important to the sheep industry. Chinese Merino (Xinjiang type) (CMXT), bred in 1985, is an excellent wool sheep breed in China, with characteristics of large body size, high density, white hair, and wool fineness between 60 and 70 count.
Hair and wool, produced from follicles, are skin appendages unique to mammals that are characterized by periodic regrowth [2]. Keratins (KRTs) and keratin-associated proteins (KAPs) are a large heterogeneous group of proteins that make up about 90% of the wool fiber [3]. The primary function of keratins is to protect epithelial cells from mechanical and non-mechanical stresses that can lead to cell death. Other emerging functions include roles in cell signaling, stress response, and apoptosis, as well as unique roles that are keratin specific and tissue specific [4]. The fibers are comprised primarily of keratin proteins, which are encoded by KRT and KAPs [5]. Although many of the keratin proteins are homologous [6], there is variation at the DNA level. Genetic variation in the keratin and keratin-associated protein (KRTAP) genes has been reported in many studies [7–10], with some authors suggesting that this variation contributes to phenotypic differences in wool [11]. This genetic diversity could have an impact on the structure of wool fiber; therefore, variation in these genes, and the proteins they encode could underpin variation in wool traits.
Keratin intermediate filament (KRT-IF) is composed of type I and II keratins; a labeled keratin intermediate filament type II (KIFII) gene was introduced into sheep and the wool quality obtained from transgenic sheep was significantly improved, with the wool fiber exhibiting high gloss and significant flexibility [12]. The quantitative trait locus affecting wool strength in Romney sheep is located on chromosome 11, and is linked to KRT1.2, B2A, and B2C (high-sulphur wool matrix protein B2A and B2C) [13]. The wool follicle genes as KRT31, KRT33a, KRT35, KRT38, KRT81, KRT83, KRT85, and KRT86 expressed in sheep keratin gene reported by [14–17]. In human hair, the keratin KRT38 gene is distributed in the cortex cells of the cortex layer, which is composed of microfibrils and macrofibrils of hair fibers [18]. The K2.9, K2.11, K2.10, and K2.12 genes are the first keratin genes to be expressed, and their transcription products are expressed in the upper part of the hair bulb [19].
The main objective of this study was to analyze the possible polymorphisms in 17 primer fragments of six candidate genes of the keratin family, including KRT27 (exons 3 and 8, introns 2 and 5), KRT31 (intron 2), KRT36 (exons 1, 2, and 3), KRT38 (exon 3, introns 2 and 7), KRT81 (exons 1 and 2, introns 1 and 3), KRT85 (exon 3, intron 2) in CMXT using the polymerase chain reaction-based single-strand conformation polymorphism (PCR-SSCP) and DNA sequencing methods. Association between the identified polymorphism sites, as well as the combined genotypes, with wool traits (staple length, wool crimps score, number of crimps, wool fineness, body size, live weight after shearing, greasy weight, fiber diameter, and coefficient of variation) were tested by least-square analysis. These findings provide a scientific basis for the improvement of wool traits through marker-assisted selection (MAS).
Genomic DNA samples were obtained from 418 healthy CMXT, which were reared at Gonaisi Fine Wool Sheep Breeding Farm, Xinyuan county, (Latitude 43°03′–43°41′N, Longitude 82°28′–84°56′E), Xinjiang Province, China. These 1-year-old sheep were from different groups, but were subjected to uniform feed and management. Genomic DNA was extracted from blood samples using the phenol-chloroform extraction method and stored at −80°C. Records of wool traits (staple length, wool crimps score, number of crimps, wool fineness, body size, live weight after shearing, greasy weight, fiber diameter, and coefficient of variation) were collected for association analysis.
On the basis of the sequences of the KRT27, KRT31, KRT36, KRT38, KRT81, and KRT85 genes obtained from the National Center for Biotechnology Information GenBank database (GenBank accession numbers AC_000176[521074], AC_ 000176[539597], AC_000176[520668], NC_00176[515000], AC_000162[540204], AC_000162[528459], respectively), 17 pairs of PCR primers (P1–P17, Table 1) were designed to amplify different PCR products including exon3, exon8, intervening sequence 2 (IVS2), IVS5 of KRT27; IVS2 of KRT31; exon2, exon3 of KRT36; exon3, exon7, IVS2 of KRT38; exon1, exon2, IVS1, IVS3 of KRT81; and exon3, IVS2 of KRT85. The 25-μL PCR amplification volume contained: 0.5 μM each of forward and reverse primer, 1× buffer (including 1.5 mM MgCl2), 200 μM dNTPs, 0.625 units of Taq DNA polymerase (MBI, Fermantas, Vilnius, Lithuania), and 50 to 100 ng genomic DNA as a template. The cycling protocol included 5 min at 95°C, 35 cycles of 94°C for 30 s, annealing at 56.5°C to 57.5°C corresponding to 17 different primer pairs for 30 s, and 72°C for 60 s, with a final extension at 72°C for 5 min.
The SSCP method was used to scan for mutations within the amplified regions. All PCR amplifications were subjected to SSCP analysis. Aliquots of 10-μL PCR products were mixed with a 10-μL denaturing solution (95% formamide, 25 mM ethylene diamine tetraacetic acid (EDTA), 0.025% xylene-cyanole, and 0.025% bromophenol blue), heated for 10 min at 98°C, and chilled on ice immediately. Denatured DNA was subjected to 10% non-polyacrylamide gel electrophoresis analysis run with 1× Tris-borate-EDTA (TBE) buffer for 10 to 18 h at a constant temperature of 4°C after a pre-run at 300 V for 30 min and under a constant voltage of 180 V. The gel was stained with 0.1% silver nitrate [20] and visualized with 2% NaOH solution (containing 0.1% formaldehyde). Following the detection of polymorphisms, PCR products from different electrophoresis patterns were sequenced by the Shanghai Sangon Biological Engineering Technology Services Co., LTD (Songjiang, Shanghai, China). The sequences were analyzed using DNAMAN 5.2.2 software.
Based on the genotypes of 17 KRT fragments in the analyzed population, genotype frequencies, allele frequencies, and Hardy–Weinberg equilibrium were calculated. The population genetic indices, such as He (gene heterozygosity), Ho (gene homozygosity; Ho+He = 1), and Ne (effective allele numbers; reciprocal of homozygosity) were calculated using the PopGene software (version 3.2, University of Alberta, Edmonton, AB, Canada). The polymorphic information content (PIC) was calculated using the method described by [21]. A chi-square test was applied to identify statistical significance, which was performed using SAS 8.1 software (SAS Inc., Cary, NC, USA) [22]. Associations between different genotypes and wool traits were analyzed by analysis of variance (SAS 8.1 software general linear model procedure) using the following model:
Where, Yijn is the observed value of the trait, μ is the overall population mean, gi is the gene effect, qj is the group effect, and eijn is the random error.
Associations between the different genotypes in the polymorphic fragments and wool traits were analyzed using SAS 8.1 software (SAS Inc., USA). Genetic analysis of the combined genotypes effect model was as follows:
Where, Yijn is the observed value of the trait, μ is the overall population mean, gI is the KRT36 gene effect, gII is the KRT38 gene effect, gIII is the KRT85 gene effect, qj is the group effect, and eijn is the random error.
In this study, PCR-SSCP and DNA sequencing methods were used to identify polymorphisms in the sheep KRT27, KRT31, KRT36, KRT38, KRT81, and KRT85 genes. Only one SSCP genotype was identified in the P1, P2, P3, and P4 fragments of KRT27, P6 and P7 fragments of KRT36, P9 fragment of KRT38, P12, P13, P14, and P15 fragments of KRT81, and P17 fragment of KRT85.
In the P5 fragment of KRT31 gene, two unique SSCP genotypes were observed (named AA and AB, Figure 1A). Comparison with the nucleotide sequence of the cattle KRT31 gene (GenBank Accession number AC_000176 [521074]) revealed the presence of a mutation: IVS2+50–52insG (Figure 2A). The frequencies of genotypes AA and AB were 0.40 and 0.60, respectively. Correspondingly, the frequencies of alleles A and B were 0.70 and 0.30, respectively, the analyzed population was in Hardy–Weinberg equilibrium (p>0.05).
In the P8 fragment of KRT36 gene, three unique SSCP banding patterns were observed (named CC, CD, and DD, Figure 1B). Based on the nucleotide sequence of the cattle KRT36 gene (GenBank Accession number AC_000176 [520668]), three mutations were detected in the P8 fragment. Among these mutations, EX3_62delA showed a silent mutation, EX3_ 63T/A showed a missense mutation leading to the changes Se→His, and EX3_83A/T showed a nonsense mutation (Figure 2B). The frequencies of genotypes CC, CD, and DD were 0.23, 0.21, and 0.56, respectively. Correspondingly, the frequencies of alleles C and D were 0.34 and 0.66, respectively, in the analyzed population, with Hardy–Weinberg equilibrium (p>0.05).
In the P10 and P11 fragments of KRT38 gene, three SSCP genotypes were identified (named EE, EF, and FF, Figure 1C, 1D). In the P10 fragment, the amplified polymorphic DNA fragments were sequenced and two mutations (IVS2+85G/A and IVS2+95C/T) were revealed. The frequencies of alleles E and F were 0.58 and 0.42, respectively, in the population, with Hardy–Weinberg equilibrium (p>0.05) (Figure 2C). Eleven mutations were detected in the P11 fragment. Among all mutation sites, EX7_93T/C, EX7_97A/C, EX7_112A/T, and EX7_ 126G/T showed missense mutations leading to the changes Ile→Thr, Glu→Asp, Gly→Ala, and Ala→Ser (Figure 2D). EX7_ 111G/C, EX7_113G/A, EX7_117C/A, EX7_118T/A, EX7_ 119T/C, EX7_120G/A, and EX7_121delC were silent mutations. The frequencies of alleles G and H were 0.59 and 0.41, respectively, and the analyzed population was in Hardy–Weinberg equilibrium (p>0.05).
In the P16 fragment of KRT85 gene, three SSCP banding patterns were found, named II, IJ, and JJ, respectively, Figure 1E. In order to better characterize genetic variation of the P16 fragment within the sheep KRT85 gene, the amplified polymorphic DNA fragments were sequenced and three mutations (a silent mutation EX3_49delA, a nonsense mutation EX3_ 210G/A, and a same-sense mutation EX3_354G/A), were detected (Figure 2E). The frequencies of alleles I and J were 0.70 and 0.30, respectively, in the population, with Hardy–Weinberg equilibrium (p>0.05).
Using the PopGene software (version 3.2) and the Botstein method, the population genetic indices (Ho, He, Ne, and PIC) of six KRT genes were estimated and are shown in Table 2. Hence, values for Ho varied from 0.51 to 0.58; He varied from 0.42 to 0.49; and Ne varied from 1.72 to 1.95. According to the classification of PIC (low polymorphism if PIC<0.25, medium polymorphism if PIC 0.25<PIC<0.5, and high polymorphism if PIC>0.5), the analyzed population possessed moderate genetic diversity at the five fragments, which are suitable for use as genetic markers in molecular breeding [23]. Genetic diversity is essential for species preservation and for improvement by selective breeding.
We analyzed the associations of 17 primer fragments in six KRT genes with wool traits in CMXT (n = 418). The results of the association analyses between single markers and wool traits are shown in Table 3. As no polymorphisms were found in the P1, P2, P3, P4, P6, P7, P9, P12, P13, P14, P15, and P17 fragments, these were not considered in the association analysis. However, a statistically significant association between two genotypes of the P5 fragment and fiber diameter was found in the analyzed population. Individuals with the AA genotype showed significantly (p<0.05) greater fiber diameter than those with the AB genotype. Statistically significant associations between three genotypes of the P8 fragment with wool fineness score and fiber diameter were also found in the analyzed population. Individuals with the CC genotype showed significantly (p<0.05) higher wool fineness score than those with the CD and DD genotypes.
No significant association (p>0.05) was observed between wool traits and different genotypes of the P10 fragment; therefore, these results indicate that this fragment might contain a neutral mutation. Moreover, a significant association between three genotypes of the P11 fragment and number of crimps was found in the analyzed population. Compared to the GH and HH genotypes of the P11 fragment, the GG genotype resulted in a significantly (p<0.05) higher number of crimps. Finally, statistical analysis showed that individuals in the analyzed population with the JJ genotype had a significantly (p<0.05) greater wool crimps score, body size, and fiber diameter than those with the IJ and II genotypes at fragment P16. Therefore, the sheep KRT31, KRT36, KRT38, and KRT85 genes had significant effects on wool traits, suggesting these are potential candidate genes for wool traits in MAS.
Many studies have investigated the association between gene polymorphisms and the wool traits; however, few have studied the effects of combined genotypes. Considering that intragenic allele interactions might exist in genes [24], the associations between combinations of genotypes and wool traits in CMXT were also analyzed (Table 4). Combined genotypes indeed had more profound impacts than the individual genotypes [25,26]. Based on the analysis of the combined genotypes of KRT36, KRT38, and KRT85, a total of 26 superior combined genotypes were found in the analyzed population, among which the combined genotype DD-GG-II was dominant. Individuals with the combined genotypes CC-GG-II had longer staple length, those with the combined genotypes CC-HH-IJ had a higher wool crimps score and number of crimps, individuals with combined genotypes CC-HH-JJ had higher wool fineness, individuals with combined genotypes DD-HH-JJ had higher body size, individuals with combined genotypes CC-GH-IJ had greater live weight after shearing, individuals with combined genotypes CC-GH-JJ had higher greasy weight, fiber diameter, and coefficient of variation, Therefore, these combined genotypes represent the optimal combinations, suggesting that KRT36, KRT38, and KRT85 should be used as candidate genes for sheep breeding.
In recent years, there have been a few domestic studies on sheep and goat KRT genes, and more reports on the cloning and expression of these genes. For example, goat keratin intermediate-filaments type I (KIFI) genes have been cloned, and homology of its exon 1 encoding 116 amino acids, with that of sheep, cattle, horse, and mouse was 98%, 90%, 88%, and 84%, respectively. The role of the KIFI gene in sheep and goat needs to be further investigated [27]. A full-length cDNA library was constructed from the skin tissue of Xinji fine wool sheep [28], and the expressed sequence tags showed that the KRT27 gene may affect the performance of wool traits. Using RT-PCR to detect differences in the expression of the KRT26 gene in the skin tissue of superfine and fine merino sheep, revealed that the level of KRT26 expression was lower than that in fine wool sheep [29].
In the present study, PCR-SSCP and genomic DNA sequencing methods were used to screen for genetic variation in different exons and their flanking regions within sheep KRT27, KRT31, KRT36, KRT38, KRT81, and KRT85 genes. The results revealed 20 mutation sites, IVS2+50–52insG, EX3_62delA, IVS2+85G/A, IVS2+95C/T, EX7_113G/A, EX7_117C/A, EX7 _118T/A, EX7_119T/C, EX7_120G/A, EX7_121delC, EX3_ 49delA, and EX7_111G/C, which showed silent mutations; among all mutations, EX7_93T/C, EX7_97A/C, EX7_112A/T, EX7_126G/T, and EX3_63T/A, showed missense mutations resulting in Ile→Thr, Glu→Asp, Gly→Ala, Ala→Ser, and Se→His; nonsense mutations EX3_83A/T and EX3_210G/A; and a same-sense mutation EX3_354G/A. An association between different genotypes of KRT31, KRT36, KRT38, and KRT85 and wool traits in CMXT was established in this study. These data strongly suggest that gene polymorphisms may represent genetic markers that could be used for the breeding of new sheep breeds. Results of the combined genotypes analysis indicated that a group of six combined genotypes for CMXT polymorphisms led to the appearance of better wool traits, including staple length in the CC-GG-II genotype, wool crimps score and number of crimps in the CC-HH-IJ genotype, wool fineness in the CC-HH-JJ genotype, body size in the DD-HH-JJ genotype, live weight after shearing in the CC-GH-IJ genotype, and greasy weight, fiber diameter, and coefficient of variation in the CC-GH-JJ genotype.
There have been several studies on polymorphisms in the KAP gene family and their association with wool traits in sheep and goat, and a few studies related to polymorphisms in the KRT gene family and their association with wool traits in sheep. The previous reports showed that the Hu sheep population possessed moderate polymorphisms at the KRT1.2 gene, while the other four analyzed populations possessed low levels of polymorphisms, and five sheep populations reached genetic equilibrium. Analysis of type I IRS keratin genes, KRT25, KRT26, KRT27, and KRT28, in the neck and groin of wool sheep showed that the expression of type I IRS keratin genes is closely related to the control of wool density in specific parts, and the whole wool development cycle [30]. Moreover, association analysis of KRT-IF35 gene polymorphisms and wool traits in Gansu Alpine Fine-wool sheep showed that the KRT-IF35 gene was associated with wool fiber diameter and coefficient of variation [31].
In conclusion, 20 mutation sites extended the characterization of genetic variation in the KRT31, KRT36, KRT38, and KRT85 genes. Additionally, we demonstrated significant associations between the sheep KRT31, KRT36, KRT38, and KRT85 genes and wool traits. Moreover, based on the findings of combined genotype analysis, the CC-GG-II, CC-HH-IJ, CC-HH-JJ, DD-HH-JJ, CC-GH-IJ, and CC-GH-JJ combined genotypes for CMXT could be used as molecular markers to identify superior wool traits, and KRT31, KRT36, KRT38, and KRT85 are potential candidate genes for wool traits. However, the present study presents preliminary results and further investigations are essential.
ACKNOWLEDGMENTS
This work was supported by the National Modern Agriculture Industrial System Project (Grant No. CARS-40), the National Natural Science Foundation of P.R. China (Grant No. 31360543, 31460593), and the Xinjiang Uygur Autonomous Region science and technology major projects in the 12th five-year plan (Grant No. 201230116-6).
Figure 1
Polymerase chain reaction-based single-strand conformation polymorphism (PCR-SSCP) genotypes of the keratin genes in Chinese Merino (Xinjiang type) sheep. A, B, C, D, and E represent the PCR-SSCP genotypes of P5, P8, P10, P11, and P16 primer pairs, respectively.
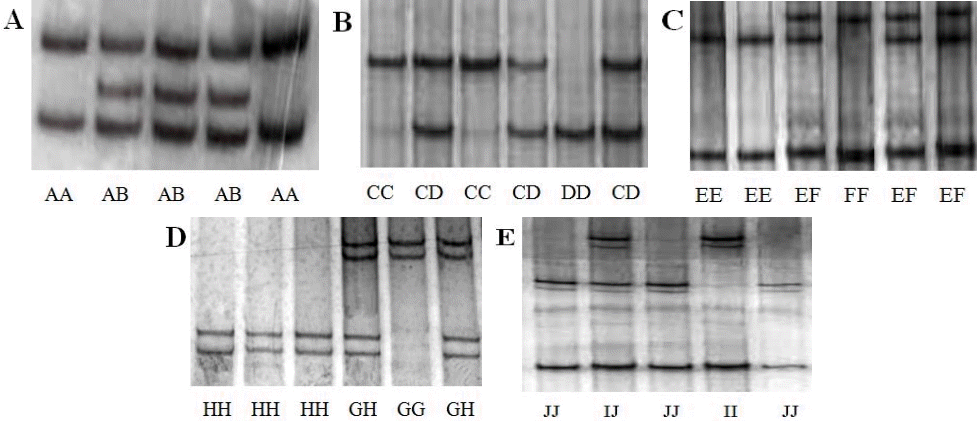
Figure 2
Sequence alignment and DNA sequencing maps of sheep keratin (KRT) genes. A, B, C, D, and E represent the polymerase chain reaction-based single-strand conformation polymorphism (PCR-SSCP) genotypes of P5, P8, P10, P11, and P16 fragments within sheep KRT-31, KRT-36, KRT-38, and KRT-85 genes.

Table 1
Information on the primer sequences within six keratin (KRT) genes in Chinese Merino (Xinjiang type) sheep
Table 2
Genotypic and allelic frequencies (%), value of χ2 test, and diversity parameters of the KRT31, KRT36, KRT38, and KRT85 genes in Chinese Merino (Xinjiang type) sheep1)
Table 3
Association between different genotypes at five primer pairs fragments of KRT31, KRT36, KRE38, and KRT85 genes with wool traits in Chinese Merino (Xinjiang type) sheep (mean±standard error)
| Wool traits | KRT31 (P5) genotypes | KRT36 (P8) genotypes | KRT38 (P10) genotypes | KRT38 (P11) genotypes | KRT85 (P16) genotypes | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
||||||||||
| AA | AB | CC | CD | DD | EE | EF | FF | GG | GH | HH | II | IJ | JJ | |
| SL | 10.869± 0.146a(238) | 10.827± 0.159a(154) | 10.801± 0.105a(92) | 10.891± 0.107a(85) | 10.806± 0.071a(221) | 10.846± 0.081a(178) | 10.841± 0.103a(96) | 10.786± 0.094a(110) | 10.867± 0.775a(177) | 10.718± 0.104a(96) | 10.859± 0.098a(109) | 10.800± 0.072a(207) | 10.749± 0.091a(122) | 10.954± 0.137a(51) |
| WCS | 4.451± 0.0281a(238) | 4.473± 0.035a(154) | 4.502± 0.043a(92) | 4.444± 0.044a(85) | 4.445± 0.029a(221) | 4.433± 0.034a(178) | 4.507± 0.044a(96) | 4.452± 0.039a(110) | 4.475± 0.024a(177) | 4.477± 0.032a(96) | 4.464± 0.031a(109) | 4.486± 0.031ab(207) | 4.385± 0.039b(122) | 4.517± 0.058a(51) |
| NC | 12.321± 0.162a(238) | 11.853± 0.204a(154) | 12.094± 0.259a(89) | 12.071± 0.263a(83) | 12.202± 0.175a(216) | 12.321± 0.198a(173) | 12.136± 0.252a(95) | 11.987± 0.232a(106) | 11.859± 0.187ab(173) | 12.288± 0.562ab(91) | 12.522± 0.238a(106) | 12.188± 0.178a(205) | 12.294± 0.227a(119) | 11.856± 0.332a(52) |
| WF | 68.260± 0.281a(238) | 67.590± 0.352a(154) | 68.556± 0.438a(92) | 68.175± 0.446ab(85) | 67.713± 0.298b(221) | 68.267± 0.338a(178) | 68.185± 0.434a(96) | 67.643± 0.396a(110) | 67.944± 0.325a(177) | 68.431± 0.436a(96) | 67.756± 0.410a(109) | 68.025± 0.308a(207) | 68.110± 0.390a(122) | 67.729± 0.584a(51) |
| BS | 4.263± 0.137a(238) | 4.514± 0.171a(154) | 4.337± 0.214a(85) | 4.207± 0.217a(92) | 4.428± 0.145a(221) | 4.244± 0.165a(178) | 4.690± 0.212a(75) | 4.237± 0.192a(110) | 4.463± 0.160a(177) | 4.321± 0.214a(96) | 4.234± 0.201a(109) | 4.226± 0.149b(207) | 4.319± 0.189b(122) | 4.997± 0.283a(51) |
| LWAS | 36.262± 0.229a(238) | 36.414± 0.288a(154) | 36.596± 0.362a(92) | 36.401± 0.368a(85) | 35.175± 0.246a(221) | 36.441± 0.272a(178) | 36.184± 0.349a(96) | 36.108± 0.318a(110) | 36.293± 0.268a(177) | 36.479± 0.361a(96) | 36.328± 0.339a(109) | 36.230± 0.250a(207) | 36.318± 0.317a(122) | 36.382± 0.475a(51) |
| GW | 3.556± 0.041a(237) | 3.578± 0.052a(150) | 3.586± 0.065a(90) | 3.637± 0.065a(84) | 3.529± 0.0434a(221) | 3.527± 0.062a(176) | 3.581± 0.062a(94) | 3.585± 0.057a(109) | 3.606± 0.047a(174) | 3.529± 0.064a(95) | 3.544± 0.059a(108) | 3.540± 0.045a(205) | 3.609± 0.056a(121) | 3.585± 0.085a(49) |
| FD | 18.601± 0.119a(236) | 18.217± 0.149b(152) | 18.785± 0.188a(89) | 18.316± 0.192b(83) | 18.381± 0.127ab(216) | 18.528± 0.144a(173) | 18.309± 0.183a(95) | 18.419± 0.168a(106) | 18.545± 0.139a(176) | 18.509± 0.190a(91) | 18.322± 0.177a(106) | 18.519± 0.128a(205) | 18.536± 0.164a(119) | 18.985± 0.241b(52) |
| CV | 20.865± 0.190a(236) | 20.932± 0.239a(152) | 21.343± 0.300a(89) | 21.121± 0.304a(83) | 20.121± 0.203a(216) | 20.905± 0.232a(173) | 21.202± 0.295a(95) | 20.673± 0.272a(108) | 20.922± 0.220a(176) | 21.036± 0.301a(91) | 20.687± 0.279a(106) | 20.903± 0.206a(205) | 20.263± 0.386a(119) | 20.737± 0.386a(52) |
Table 4
Associations of the three combined diplotypes of the KRT36, KRT38(P11), and KRT85 genes with wool traits in Chinese Merino (Xinjiang type) sheep
| Genotypes | GF | SL | WCS | NC | WF | BS | LWAS | GW | FD | CV |
|---|---|---|---|---|---|---|---|---|---|---|
| CC-GG-II(25) | 0.07 | 11.85±0.19abc | 4.55±0.06a | 12.91±0.47abcd | 69.11±0.84ab | 4.23±0.10abc | 35.35±0.69ab | 3.69±0.12abcd | 16.89±0.24bcd | 21.09±0.56bc |
| CC-GG-IJ(8) | 0.02 | 10.06±0.39abc | 4.57±0.11a | 12.23±0.84abcd | 68.58±1.46ab | 4.13±0.18abc | 35.85±1.21ab | 3.11±0.21cd | 17.16±0.42bcd | 20.88±0.99bc |
| CC-GG-JJ(6) | 0.02 | 10.52±0.40abc | 4.54±0.12a | 11.71±0.97abcd | 67.97±1.69ab | 3.96±0.20abc | 35.45±1.40ab | 3.57±0.24bcd | 16.52±0.49bcd | 21.25±1.15bc |
| CC-GH-II(6) | 0.02 | 10.67±0.40abc | 4.65±0.12a | 14.08±0.97ab | 71.64±1.71a | 4.25±0.21abc | 36.18±1.41ab | 3.44±0.25bcd | 16.84±0.49bcd | 21.61±1.16abc |
| CC-GH-IJ(10) | 0.03 | 10.79±0.31abc | 4.35±0.09a | 11.38±0.75abcd | 67.95±1.31b | 4.39±0.16abc | 37.49±1.09ab | 3.79±0.19ab | 16.43±0.38cd | 21.82±0.89abc |
| CC-GH-JJ(3) | 0.01 | 10.24±0.56bc | 4.54±0.17a | 11.08±1.36bcd | 67.18±2.38ab | 4.01±0.29bc | 33.38±1.97c | 4.23±0.34a | 16.15±0.68d | 23.62±1.61a |
| CC-HH-II(10) | 0.03 | 10.51±0.31abc | 4.58±0.09a | 13.06±0.75abcd | 70.01±1.31ab | 4.12±0.16abc | 35.64±1.08bc | 3.61±0.19abcd | 16.31±0.38d | 20.42±0.89bc |
| CC-HH-IJ(11) | 0.03 | 10.71±0.29abc | 4.68±0.09a | 14.12±0.72a | 69.54±1.26ab | 4.34±0.15abc | 36.41±1.04ab | 3.40±0.18bcd | 16.72±0.36bcd | 20.35±0.86bc |
| CC-HH-JJ(5) | 0.01 | 10.16±0.44c | 4.46±0.14a | 13.03±1.07abcd | 70.22±1.87ab | 4.11±0.23abc | 34.68±1.55bc | 3.07±0.27bcd | 16.27±0.54d | 21.31±1.27abc |
| CD-GG-II(23) | 0.07 | 10.66±0.20abc | 4.49±0.06a | 11.48±0.49abcd | 68.40±0.86ab | 4.39±0.10abc | 36.14±0.71ab | 3.61±0.12abcd | 17.48±0.25bcd | 20.99±0.58abc |
| CD-GG-IJ(7) | 0.02 | 9.84±0.37c | 4.38±0.11a | 11.93±0.89abcd | 68.12±1.56ab | 4.04±0.20abc | 35.84±1.29ab | 3.62±0.24abcd | 17.78±0.45bc | 21.32±1.06abc |
| CD-GG-JJ(5) | 0.01 | 11.09±0.43ab | 4.48±0.13a | 10.92±1.05bcd | 67.56±1.84b | 4.18±0.22abc | 36.17±1.52ab | 3.45±0.29bcd | 17.79±0.53b | 19.04±1.25bc |
| CD-GH-II(13) | 0.04 | 10.25±0.27abc | 4.43±0.08a | 11.39±0.65abcd | 68.52±1.14ab | 4.04±0.14c | 35.49±0.95abc | 3.29±0.17cd | 17.44±0.33bcd | 19.48±0.78bc |
| CD-GH-IJ(10) | 0.03 | 10.89±0.31abc | 4.52±0.09a | 12.82±0.75abcd | 67.58±1.32b | 4.18±0.16abc | 35.99±1.09ab | 3.77±0.19abcd | 17.78±0.38b | 19.83±0.89bc |
| CD-GH-JJ(1) | 0.02 | 9.85±0.32abc | 4.40±0.08a | 11.79±0.64abcd | 65.48±1.10b | 4.14±0.13abc | 35.83±0.69abc | 3.28±0.21abcd | 17.56±0.30b | 19.72±0.77bc |
| CD-HH-II(9) | 0.03 | 10.78±0.33abc | 4.43±0.09a | 11.87±0.79abcd | 62.49±1.38b | 4.12±0.16abc | 36.53±1.14ab | 3.56±0.20bcd | 17.84±0.40b | 21.71±0.94ab |
| CD-HH-IJ(6) | 0.02 | 10.86±0.39abc | 3.74±0.12b | 12.13±0.96abcd | 67.41±1.69b | 4.01±0.20abc | 35.04±1.39abc | 3.48±0.24abcd | 17.56±0.49bcd | 20.12±1.15bc |
| CD-HH-JJ(5) | 0.01 | 10.85±0.43abc | 4.42±0.13a | 13.74±1.05abc | 66.31±1.85b | 4.53±0.22a | 36.23±1.53ab | 3.83±0.27abc | 17.75±0.53b | 21.48±1.25abc |
| DD-GG-II(58) | 0.19 | 11.02±0.13abc | 4.45±0.04a | 11.78±0.32abcd | 67.49±0.55b | 4.31±0.07abc | 36.61±0.46ab | 3.57±0.08abcd | 19.59±0.16a | 20.93±0.38bc |
| DD-GG-IJ(30) | 0.09 | 10.74±0.18abc | 4.45±0.06a | 11.79±0.43abcd | 68.05±0.76b | 4.43±0.09abc | 36.03±0.63ab | 3.69±0.12abcd | 19.38±0.22a | 21.19±0.51abc |
| DD-GG-JJ(9) | 0.03 | 11.37±0.32a | 4.55±0.10a | 11.83±0.78abcd | 67.36±1.38b | 4.18±0.17abc | 36.23±1.14ab | 3.59±0.19abcd | 19.09±0.39a | 20.37±0.93bc |
| DD-GH-II(24) | 0.07 | 10.79±0.49abc | 4.49±0.06a | 12.75±0.49abcd | 68.31±0.86ab | 4.23±0.10abc | 36.42±0.71ab | 3.43±0.12bcd | 19.98±0.25a | 21.18±0.58abc |
| DD-GH-IJ(17) | 0.05 | 10.83±0.24abc | 4.44±0.07a | 12.53±0.63abcd | 67.86±1.01b | 4.40±0.12abc | 36.65±0.84ab | 3.68±0.15abcd | 20.13±0.32a | 21.09±0.76abc |
| DD-GH-JJ(9) | 0.03 | 10.82±0.33abc | 4.58±0.09a | 12.51±0.84abcd | 68.42±1.38ab | 4.28±0.17abc | 36.59±1.14ab | 3.42±0.21bcd | 19.23±0.42a | 21.71±0.99abc |
| DD-HH-II(29) | 0.09 | 10.96±0.18abc | 4.48±0.06a | 12.86±0.44abcd | 67.94±0.78b | 4.24±0.09abc | 36.28±0.64ab | 3.47±0.11bcd | 19.59±0.22a | 20.84±0.53bc |
| DD-HH-IJ(19) | 0.05 | 11.09±0.23ab | 4.47±0.07a | 12.27±0.55abcd | 67.29±0.96b | 4.49±0.11abc | 37.32±0.792ab | 3.69±0.138abc | 19.96±0.27a | 21.03±0.65abc |
| DD-HH-JJ(7) | 0.02 | 10.86±0.38abc | 4.47±0.11a | 10.55±0.84bcd | 67.68±1.56b | 4.65±0.19ab | 39.18±1.289a | 3.90±0.225abc | 19.41±0.42a | 19.79±0.99abc |
REFERENCES
1. Itenge TO, Hickford J, Forrest R, Mckenzie GW, Frampton CM. Improving the quality of wool through the use of gene markers. S Afr J Anim Sci 2010; 39:219–23.

2. Yu Z, Wildermoth JE, Wallace OAM, et al. Annotation of sheep keratin intermediate filament genes and their patterns of expression. Exp Dermatol 2011; 20:582–8.


3. Mclaren RJ, Rogers GR, Davies KP, Maddox JF, Montgomery GW. Linkage mapping of wool keratin and keratin-associated protein genes in sheep. Mamm Genome 1997; 8:938–40.


4. Coulombe PA, Omary MB. ‘Hard’ and ‘soft’ principles defining the Structure, function and regulation of keratin intermediate filaments. Curr Opin Cell Biol 2002; 14:110–22.


5. Jollès P, Zahn H, Höcker H. Formation and structure of human hair. Basel, Switzerland: Birkhäuser Verlag; 2001.
6. Plowman JE. The proteomics of keratin proteins. J Chromatogr B Analyt Technol Biomed Life Sci 2007; 849:181–9.


7. Rogers GR, Hickford JGH, Bickerstaffe R. Mspl RFLP in the gene for a Type I intermediate filament wool keratin. Anim Genet 1993; 24:218

8. Rogers GR, Hickford JGH, Bickerstaffe R. Polymorphism in two genes for B2 high sulphur proteins of wool. Anim Genet 1994; 25:407–15.


9. Beh KJ, Callaghan MJ, Leish Z, et al. A genome scan for QTL affecting fleece and wool traits in Merino sheep. Wool Technol Sheep Breed 2001; 49:88–97.
10. Intengemweza TO, Forrest RH, McKenzie GW, et al. Polymorphism of the KAP1.1, KAP1.3 and K33 genes in Merino sheep. Mol Cell Probes 2007; 21:338–42.


11. Purvis IW, Franklin IR. Major genes and QTL influencing wool production and quality: a review. Genet Sel Evol 2005; 37:S97–S107.



12. Bawden CS, Sivaprasad AV, Verma PJ, Walker SK, Rogers GE. Expression of bacterial cysteine biosynthesis genes in transgenic mice and sheep: toward a new in vivo amino acid biosynthesis pathway and improved wool growth. Transgenic Res 1995; 4:87–104.


13. Powell B, Crocker L, Rogers G. Hair follicle differentiation: expression, structure and evolutionary conservation of the hair type II keratin intermediate filament gene family. Development 1992; 114:417–33.



14. Powell BC, Beltrame JS. Characterization of a hair (wool) keratin intermediate filament gene domain. J Invest Dermatol 1994; 102:171–7.


15. Rufaut NW, Pearson AJ, Nixon AJ, Wheeler TT, Wilkins RJ. Identification of differentially expressed genes during a wool follicle growth cycle induced by prolactin. J Invest Dermatol 1999; 113:865–72.


16. Yu Z, Gordon SW, Nixon AJ, et al. Expression patterns of keratin intermediate filament and keratin associated protein genes in wool follicles. Differentiation 2009; 77:307–16.


17. Liu YF, Kang XL, Yang WJ, et al. Differential expression of KRT83 regulated by the transcript factor CAP1 in Chinese Tan sheep. Gene 2017; 614:15–20.


18. Langbein L, Rogers MA, Winter H, et al. The catalog of human hair keratin. I: expression of the nine type I members in the hair follicle. J Biol Chem 1999; 274:19874–84.


19. Di J, Wang Q, Li Y; LAZHTI. The keratinis genes and its expression in the wool follicle. Grass-feeding Livest 2004; 17–19.
20. Sun HS, Anderson LL, Yu TP, et al. Neonatal Meishan pigs show POU1F1 genotype effects on plasma GH and PRL concentration. Anim Reprod Sci 2002; 69:223–37.


21. Botstein D, White RL, Skolnick M, Darvis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 1980; 32:314–31.


22. Mead R, Curnow RN. Statistical methods in agriculture and experimental biology. J Royal Stat Soc: Series A (Statistics in Society) 1994; 43:464

23. Moazamigoudarzi K. A set of 99 cattle microsatellites: characterization, synteny mapping, and polymorphism. Mamm Genome 1994; 5:288–97.


24. Boulesteix AL, Strobl C, Weidinger S, Wichmann HE, Wagenpfeil S. Multiple testing for SNP-SNP interactions. Stat Appl Genet Mol Biol 2007; 6:122–42.


25. Kang ES, Kim HJ, Nam M, et al. A novel 111/121 diplotype in the Calpain-10 gene is associated with type 2 diabetes. J Hum Genet 2006; 51:629–33.


26. Rodriguez S, Eiriksdottir G, Gaunt TR, et al.
IGF2BP1, IGF2BP2 and IGF2BP3 genotype, haplotype and genetic model studies in metabolic syndrome traits and diabetes. Growth Horm IGF Res 2010; 20:310–8.



27. Li-Juan LI, Song DR, Han FX. Cloning and sequence analysis of goat KIFI gene. China Anim Husb Vet Med 2010; 37:91–3.
28. Wu X, Tian KC, Liu WJ, et al. Construction of a long-lenth cDNA library for Xinji Fine-wool Sheep skin tissue by use of SMART technique. J Northwest A & F Univ 2010; 38:46–50.
29. Tian YZ, Jiang DI, Wei-Wei WU, et al. Expression difference of keratin 26 gene of skin tissues in superfine and fine Merino sheep. China Anim Husb Vet Med 2012; 39:31–7.
30. Juanjuan YU, Liu J, Zhao J, et al. Gene chip analysis of expression pattern of type i inner root sheath (IRS) keratin in AoHan wool sheep. Agric Sci Technol 2012; 13:1171–4.
31. Wang LP, Yang BH, Yue YJ, et al. Analysis on genetic polymorphism of KRT35 gene sequence of Gansu alpine fine-wool sheep. Guizhou Agric Sci 2010; 38:142–4.






 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print





