 |
 |
Abstract
Interferon-tau (IFNT) is thought to be the conceptus protein that signals maternal recognition of pregnancy in ruminants. We and others have observed that OCT4 expression persists in the trophectoderm of ruminants; thus, both CDX2 and OCT4 coexist during the early stages of conceptus development. The aim of this study was to examine the effect of CDX2 and OCT4 on IFNT gene transcription when evaluated with other transcription factors. Human choriocarcinoma JEG-3 cells were cotransfected with an ovine IFNT (-654-bp)-luciferase reporter (-654-IFNT-Luc) construct and several transcription factor expression plasmids. Cotransfection of the reporter construct with Cdx2, Ets2 and Jun increased transcription of -654-IFNT-Luc by about 12-fold compared with transfection of the construct alone. When cells were initially transfected with Oct4 (0 h) followed by transfection with Cdx2, Ets2 and/or Jun 24 h later, the expression of -654-IFNT-Luc was reduced to control levels. OCT4 also inhibited the stimulatory activity of CDX2 alone, but not when CDX2 was combined with JUN and/or ETS2. Thus, when combined with the other transcription factors, OCT4 exhibited little inhibitory activity towards CDX2. An inhibitor of the transcriptional coactivator CREB binding protein (CREBBP), 12S E1A, reduced CDX2/ETS2/JUN stimulated -654-IFNT-Luc expression by about 40%, indicating that the formation of an appropriate transcription factor complex is required for maximum expression. In conclusion, the presence of OCT4 may initially minimize IFNT expression; however, as elongation proceeds, the increasing expression of CDX2 and formation of the transcription complex leads to greatly increased IFNT expression, resulting in pregnancy establishment in ruminants.
In farm animals, up to 45% of conceptuses fail to implant to the uterine endometrium where embryonic mortality occurs most frequently during the first to third week period after conception (Roberts et al., 1992). During this period, conceptuses of ruminants such as cow and sheep go through blastocyst formation, hatching, migration, elongation, apposition, attachment and limited invasion to the uterine epithelium, and the initiation of placental formation. For these processes, an outer layer of the blastocyst, trophectoderm, goes through extensive cell proliferation as well as differentiation. These changes in the trophectoderm are regulated through numerous gene expressions in a temporal and spatial manner both at the blastocyst and uterus. The regulation of these changes become critical, if a pregnancy is to be established.
Gene expressions in the formation of blastocysts have been extensively studied in mice. As blastocyst development proceeds, the POU transcription factor, OCT4 (Pou5f1), becomes progressively restricted, first to the entire inner cell mass (ICM) and then to the epiblast (Palmieri et al., 1994; Nichols et al., 1998; Ralston and Rossant, 2005). Caudal related homeobox 2 transcription factor, CDX2, is involved in the control of cell differentiation into the trophectoderm (TE) and the intestinal epithelium in mice (Suh et al., 1994; Beck et al., 1995; Chawengsaksophak et al., 1997). OCT4 and CDX2 have an inverse relationship in early embryos, and the expression of CDX2 in pre- and early post-implantation embryos is tightly restricted to the TE lineage (Beck et al., 1995; Niwa et al., 2005). Recently, Berg and coworkers (2011) demonstrated that OCT4 transcripts coexist with CDX2 in the bovine TE and bovine OCT4 gene transfected into the mouse TE is actively transcribed. These investigators concluded that OCT4 and CDX2 do not coexist in the mouse TE because it implants to the maternal endometrium in a very short time period while in other species including humans, down-regulation of OCT4 is slow, resulting in the expression of both factors in the TE (Berg et al., 2011).
Interferon-tau (IFNT), produced by the mononucleate TE of ruminant ungulates during the peri-implantation period (Farin et al., 1989; Guillomot et al., 1990; Demmers et al., 2001), is a major protein that signals the process of maternal recognition of pregnancy (Godkin et al., 1982; Imakawa et al., 1987; Stewart et al., 1987; Roberts et al., 1992). However, a method to effectively control IFNT production and to improve pregnancy rate has not been developed despite the fact that IFNT was identified over two decades ago. Using human choriocarcinoma (JEG-3 or JAR) cells, transcription factor AP-1 (official symbol, JUN) and ETS2 binding sites at the distal and proximal promoter regions, respectively, have been shown to be effective in IFNT-reporter transactivation (Ezashi et al., 1998, 2001; Yamaguchi et al., 1999, 2001). Additionally, Ezashi et al. (2001) found that OCT4 binds to and inhibits the action of ETS2 on bovine IFNT in a heterologous over-expression system. These results suggest that the POU domain of OCT4 interacts directly with the central domain of ETS2 to act as a co-repressor. According to Ezashi et al. (2001), OCT4 expression subsides long before the initiation of conceptus elongation, when ETS2 begins to activate bovine IFNT gene transcription. In situ hybridization results suggested that OCT4 mRNA was restricted to the ICM in d 7 bovine blastocysts (Kurosaka et al., 2004; Saadeldin et al., 2012). However, further studies revealed that OCT4 expression remained until d 12 and 13, just prior to conceptus elongation (Degrelle et al., 2005), or even longer period (Berg et al., 2011).
We have previously shown that, although the proximal promoter is undoubtedly required, the upstream JUN binding site of the distal enhancer region is also necessary for ovine IFNT gene transcription (Yamaguchi et al., 1999; 2001; Matsuda et al., 2004). In addition, the formation of a protein complex between CREBBP, JUN, and ETS2 is required for IFNT gene transcription (Xu et al., 2003). We recently demonstrated that IFNT expression depends not only on the JUN binding site but also on the CDX2 binding site (Imakawa et al., 2006; Sakurai et al., 2010). Moreover, we and others have shown that the CDX2 transcription factor is expressed in ovine and bovine blastocysts during the conceptus elongation period (Degrelle et al., 2005; Imakawa et al., 2006; Berg et al., 2011). In fact, the over-expression of CDX2, JUN, and ETS2 in JEG-3 cells was very effective in increasing the degree of ovine IFNT-reporter transcription (Imakawa et al., 2006).
The high acetylation and low methylation (euchromatin) status of histone proteins has recently been shown to be required for IFNT gene transcription (Sakurai et al., 2009; 2010). More importantly, this euchromatin state is maintained through the expression of CDX2 and CREBBP. Although the involvement of OCT4 in the regulation of bovine IFNT gene transcription has been demonstrated (Ezashi et al., 2001), the relationship between CDX2, CREBBP, OCT4, and other transcription factors in IFNT gene transcription has not been carefully studied. Thus, the coexistence of OCT4 and CDX2 in TE led us to investigate a possible relationship between these and other IFNT gene-related transcription factors in sheep.
Total RNAs from conceptuses on d 15, 17, and 21 (n = 4 each) were extracted using Isogen Reagent (Nippon Gene, Toyama, Japan) according to the manufacturerŌĆÖs protocol. RNA (1 ╬╝g) extracted from each sample was subjected to reverse transcription (RT) into cDNA using oligo(dT) 12 to 18 primers and SuperScript II (Gibco BRL Life Technologies, Rockville, MD, USA) according to the manufacturerŌĆÖs instructions, and the RT products were used as templates for PCR analysis.
Levels of OCT4 and CDX2 as well as CREBBP, JUN, ETS2 and IFNT mRNAs in peri-implantation conceptuses were examined by PCR using specific primers (Table 1). The PCR mixture consisted of 1 ╬╝l of RT product, 1 ╬╝l of 10├ŚPCR buffer, 0.4 ╬╝l each of forward and reverse primers (10 pM), 0.2 ╬╝l of dNTP mixture (10 mM), 0.3 ╬╝l of MgCl2 (50 mM), 6.6 ╬╝l of ddH2O, and 0.1 ╬╝l of Taq DNA polymerase (5 U/╬╝l; Invitrogen, Carlsbad, CA, USA). PCR was performed under the following conditions: 94┬░C for 5 min, followed by 30 cycles of 94┬░C for 1 min, 57┬░C for 1 min, and 72┬░C for 1 min. The PCR product was analyzed by electrophoresis on a 1% agarose gel stained with ethidium bromide. The cDNA fragment was extracted from the agarose gel using the QIAquick Gel Extraction Kit (Qiagen, Tokyo, Japan) and then cloned into pGEM-T Easy Vector (Promega, Madison, WI, USA). Nucleotide sequences were determined by DNA sequencing (ABI-PRISM, Foster City, CA, USA). ACTB mRNA was used as an internal control (Sakurai et al., 2009).
The upstream region (between ŌłÆ654 and +51 bp) of ovine IFNT gene (IFNŽä-o10; Nephew et al., 1993; GenBank accession number: M88773) and its transcription factor binding-site mutants was amplified by PCR using specific primers (Yamaguchi et al., 1999; Matsuda et al., 2004). The PCR product was inserted into the pGL3-Basic Vector (Promega), which contained a firefly luciferase gene. All expression vectors, pSG5 (Promega)-based constructs, were driven by the SV40 promoter/enhancer. The plasmid constructs were all confirmed to contain the desired nucleotide sequences by dideoxy sequencing.
Human choriocarcinoma JEG-3 cells (HTB36; American Type Culture Collection) were cultured in DulbeccoŌĆÖs modified EagleŌĆÖs medium (DMEM; Sigma-Aldrich, St. Louis, MO) supplemented with 10% (v/v) fetal bovine serum (FBS; Sigma-Aldrich), 40 U/ml penicillin, and 40 ╬╝g/ml streptomycin under 5% CO2 at 37┬░ C. JEG-3 cells were re-plated onto 24-well plastic culture plates for 24 h and allowed to reach 60% to 70% confluence. Cells were transiently transfected with reporter constructs and/or expression plasmids using 3 ╬╝l of TransFast (cationic lipids; Promega) according to the manufacturerŌĆÖs protocol. To each well of the 24-well plates, 1 ╬╝g of total plasmid DNA was added in duplicate (12 treatments/plate, n = 4) in 200 ╬╝l DMEM. After 1 h incubation at 37┬░C, 1 ml of DMEM containing 10% FBS was added to each well, and the transfected cells were cultured for 48 h. To normalize transfection efficiency, the pRL-TK vector (containing the herpes simplex virus thymidine kinase promoter and the Renilla luciferase gene; Promega), along with each pGL3 vector, was transfected into JEG-3 cells. The ratio of pGL3 to pRL-TK was optimized at 160:1 (Imakawa et al., 2006).
For cotransfection experiments, pGL3 (800 ng), expression vector (140 ng), and pRL-TK (5 ng) plasmids were added to the medium. For cotransfection of 2 or more types of expression vectors, the ratio of the plasmids was 1:1:1 (35:35:35 ng). Throughout cotransfection studies, the total amounts of expression plasmids were kept constant by using the pSG5-based empty plasmid.
For the 0/24 h transfection experiment, the initial transfection at 0 h was performed as mentioned above. After 24 h of incubation, the medium containing 10% FBS was replaced with medium without FBS, and transfection of expression plasmid(s) was performed in the same manner. These cells were incubated for a further 24 h.
For immunoprecipitation studies, 24 ╬╝g of expression plasmids was transfected into JEG-3 cells, which had been cultured to 70% to 80% confluence. Two kinds of plasmids (12 ╬╝g each) or 4 kinds of plasmids (6 ╬╝g each) were transfected into JEG-3 cells by the aforementioned method.
Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturerŌĆÖs protocol. At 48 h of incubation following the plasmid transfection, JEG-3 cells were washed with phosphate-buffered saline (PBS) and lysed with Passive Lysis Buffer (Promega). A total of 20 ╬╝l each of the various lysates was placed into a 96-well plate and luciferase activity was measured using a luminometer (PerkinElmer, Foster City, CA, USA). The luciferase activity resulting from the various ovine IFNT-reporter constructs was normalized by dividing the luminescence of firefly luciferase by that of Renilla luciferase (Imakawa et al., 2006). Luciferase activities were expressed as fold activation relative to an appropriate control, the ovine IFNT-reporter construct without expression plasmids, within the experiment.
Nuclear proteins were extracted from JEG-3 cells (5├Ś106 cells) transfected with expression constructs according to the protocol described previously (Matsuda et al., 2004; Imakawa et al., 2006). Concentrations of nuclear extracts were determined by using the Bio-Rad Protein Assay Dye Reagent Concentrate (Bio-Rad Laboratories, Hercules, CA, USA). A total of 10 ╬╝g of nuclear extracts was loaded onto each lane and separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins in the gel were transferred onto polyvinylidene difluoride (PVDF) membrane (pore size, 0.45 ╬╝m; Millipore, Bedford, MA, USA) by electro-blotting (Matsuda et al., 2004) and then treated with rabbit polyclonal antibody against the DNA-binding domain of mouse JUN (D, 0.04 ╬╝g/ml; Santa Cruz Biotechnology, Santa Cruz, CA, USA), the C-terminus of human ETS2 (C-20, 0.04 ╬╝g/ml; Santa Cruz Biotechnology), the N-terminus of human CDX2 (ab22586, 0.04 ╬╝g/ml; Abcam, Cambridge, UK), the N-terminus of human OCT4 (N-19, 0.04 ╬╝g/ml; Santa Cruz Biotechnology), or the C-terminus of human CREBBP (C-1, 0.04 ╬╝g/ml; Santa Cruz Biotechnology). Normal rabbit IgG (Sigma-Aldrich) was used as a negative control. The membrane was treated with the secondary antibody, donkey anti-rabbit IgG conjugated with horseradish peroxidase (Amersham Pharmacia Biotech, Buckinghamshire, UK), and then detected by using an ECL western blotting detection system (Amersham Pharmacia Biotech).
For immunoprecipitation studies, mouse monoclonal antibody against human CREBBP (C-1, 20 ╬╝g; Santa Cruz Biotechnology) was incubated with 10 ╬╝l of protein A Sepharose beads (Amersham Pharmacia) at 4┬░C for 4 h (Imakawa et al., 2006; Sakurai et al., 2009; 2010). Anti-CREBBP antibody pretreated with blocking peptide was used as a negative control. The beads were pelleted and washed 3 times in PBS containing 0.1% Tween-20 (Wako Chemical Industries, Ltd., Osaka, Japan). The pellets were then mixed with 40 ╬╝g of nuclear proteins prepared from JEG-3 cells and incubated at 4┬░C overnight. The mixture was pelleted and washed 3 times in PBS containing 1% Tween-20, and the final pellets were suspended and separated by 10% SDS-PAGE. The immunoprecipitates were assayed by western blotting using rabbit polyclonal antibody against JUN (D, 0.04 ╬╝g/ml) or ETS2 (C-20, 0.04 ╬╝g/ml) (Santa Cruz Biotechnology). Normal rabbit IgG (Sigma-Aldrich) was used as a negative control.
The levels of CDX2 mRNA were highest on d 17, with expression patterns preceding those of IFNT mRNA. Strong expression of OCT4 mRNA was detected on d 15, 17, and 21 of gestation. Similar levels of transcription factor CREBBP, JUN, ETS2 mRNAs were found this time period whereas IFNT mRNA declined (Figure 1A). The efficacy of all pSG5-based expression vectors was examined by transient transfection into JEG-3 cells, from which the nuclear proteins (cell lysates) were isolated and subjected to western blot analysis. All expression constructs exhibited proteins of the expected sizes (Figure 1B).
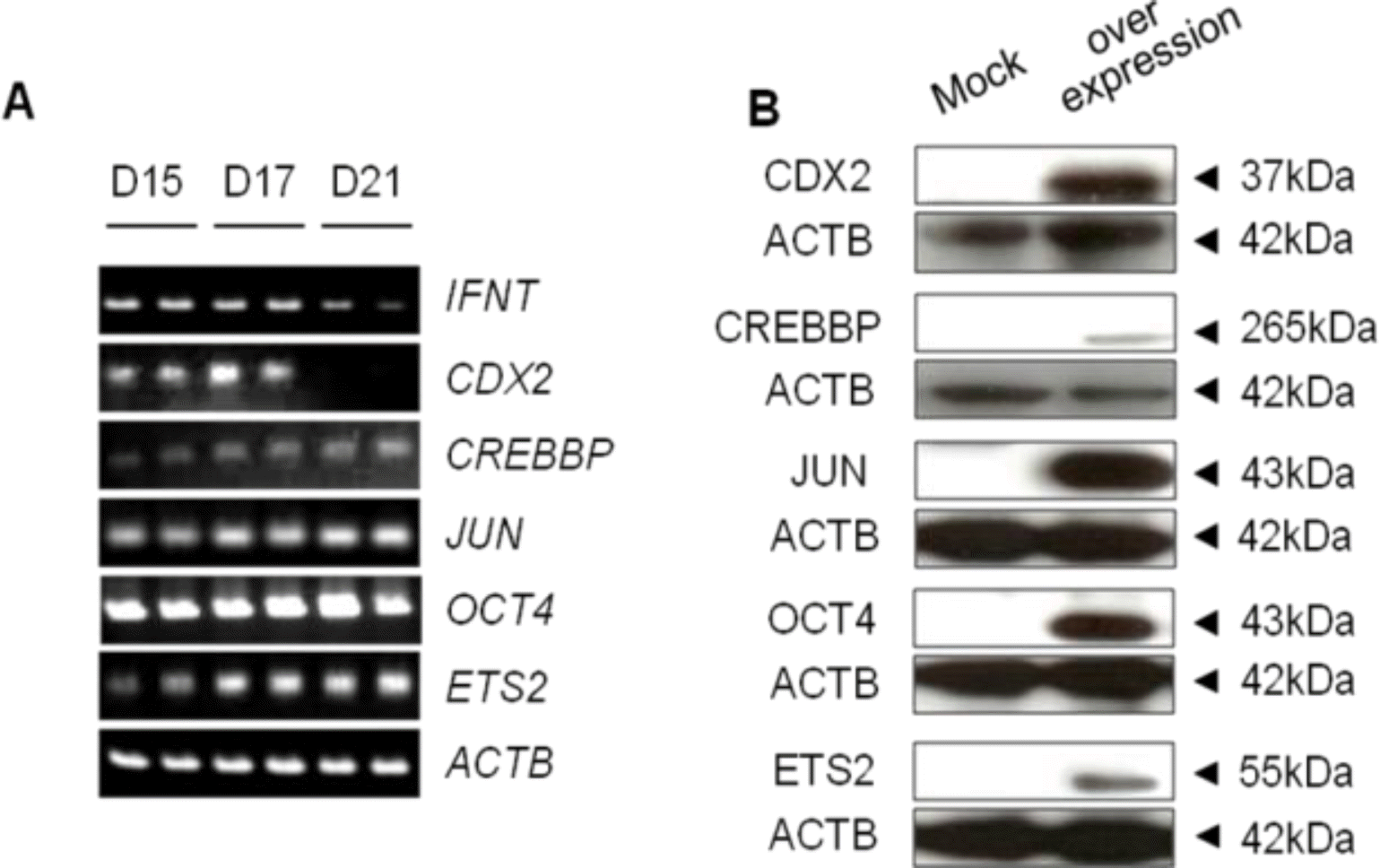
Similar to our previous study (Imakawa et al., 2006), the luciferase activity of the -654-IFNT-Luc transfected with Jun, Ets2 and Cdx2 expression plasmids was approximately 12 times that of the -654-IFNT-Luc plasmid alone (Figure 2). To investigate whether OCT4 affected ovine IFNT gene transcription, the -654-IFNT-Luc construct was transiently transfected into JEG-3 cells with Oct4 in combination with Jun, Ets2 and/or Cdx2 expression plasmids, and luciferase activities were measured. OCT4 inhibited transcription of -654-IFNT-Luc when either Jun, Ets2, Cdx2, or Jun/Ets2 together with the reporter construct was transfected into JEG-3 cells. However, OCT4 did not suppress the level of the -654-IFNT-Luc transcription when Cdx2 was cotransfected into JEG-3 cells along with Jun, Ets2, or Jun/Ets2.
To investigate whether preventing the binding of JUN, ETS2, and/or CDX2 resulted in decreased IFNT gene transcription, the upstream binding sites of the -654-IFNT-Luc reporter construct were mutated and transfected into JEG-3 cells along with the 3 transcription factors. As shown in Figure 3, mutation of any of the binding sites resulted in reduced luciferase activities. These results indicate that JUN, ETS2, and CDX2 transcription factors may form a complex (or complexes) required for ovine IFNT gene transcription, and that each factor requires a specific binding site on the upstream region of the ovine IFNT gene. In addition, cotransfection of the Oct4 expression plasmid along with Jun, Ets2 and Cdx2 did not change the level of luciferase activity, which had already been reduced due to mutation of the binding sites.
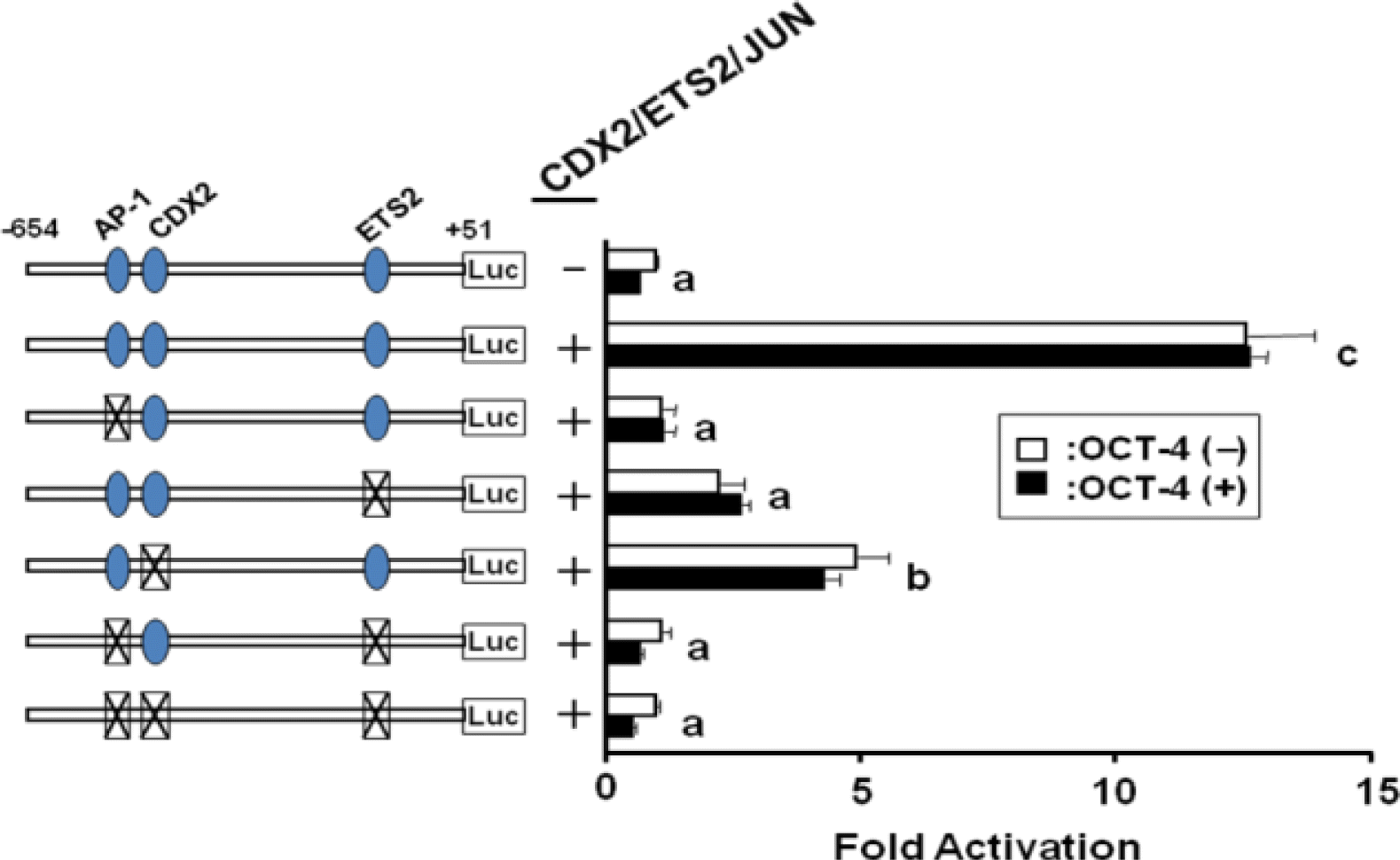
Since OCT4 over-expression concurrent with transfection of Jun, Ets2, and Cdx2 did not inhibit activity on -654-IFNT-Luc transcription, the effect of the timing of transcription factor expression on -654-IFNT-Luc transcription was examined. Experiments were performed using various combinations of transcription factor expression plasmids, which were transfected into JEG-3 cells at 0 or 24 h after the initiation of the experiment. The maximum inhibitory effect of OCT4 on ovine IFNT gene transcription was found when Oct4 alone was transfected prior to the transfection of any other transcription factor expression plasmids (0 h; Figure 4). Transfection of Oct4 at 24 h did not inhibit IFNT-Luc transcription. The results of the 0/24 h experiment were consistent with our hypothesis that substantial IFNT gene transcription requires the formation of a transcription factor complex.
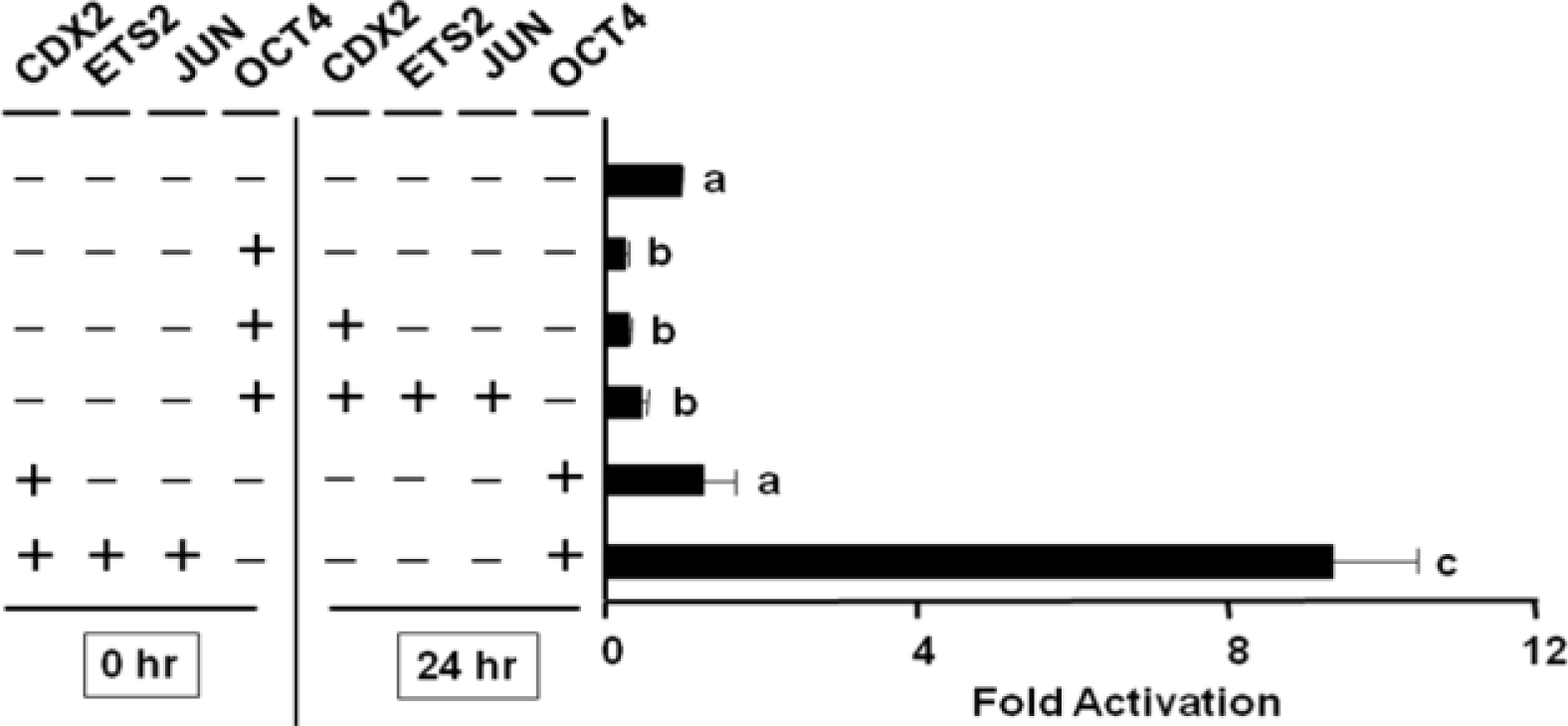
To evaluate the role of CREBBP on IFNT transcription, JEG-3 cells were cotransfected with the -654-IFNT-Luc construct along with Jun, Ets2, and/or Cdx2 expression plasmids, either with or without Oct4 or a CREBBP inhibitor, 12S E1A. No additional increase in ovine IFNT gene transcription was observed when Crebbp was transfected along with Jun, Ets2 and/or Cdx2 (Figure 5). However, co-transfection of 12S E1A reduced the level of ovine IFNT gene transcription even in the presence of CDX2, and the level of luciferase activity reduction was greater than that resulting from Oct4 cotransfection.
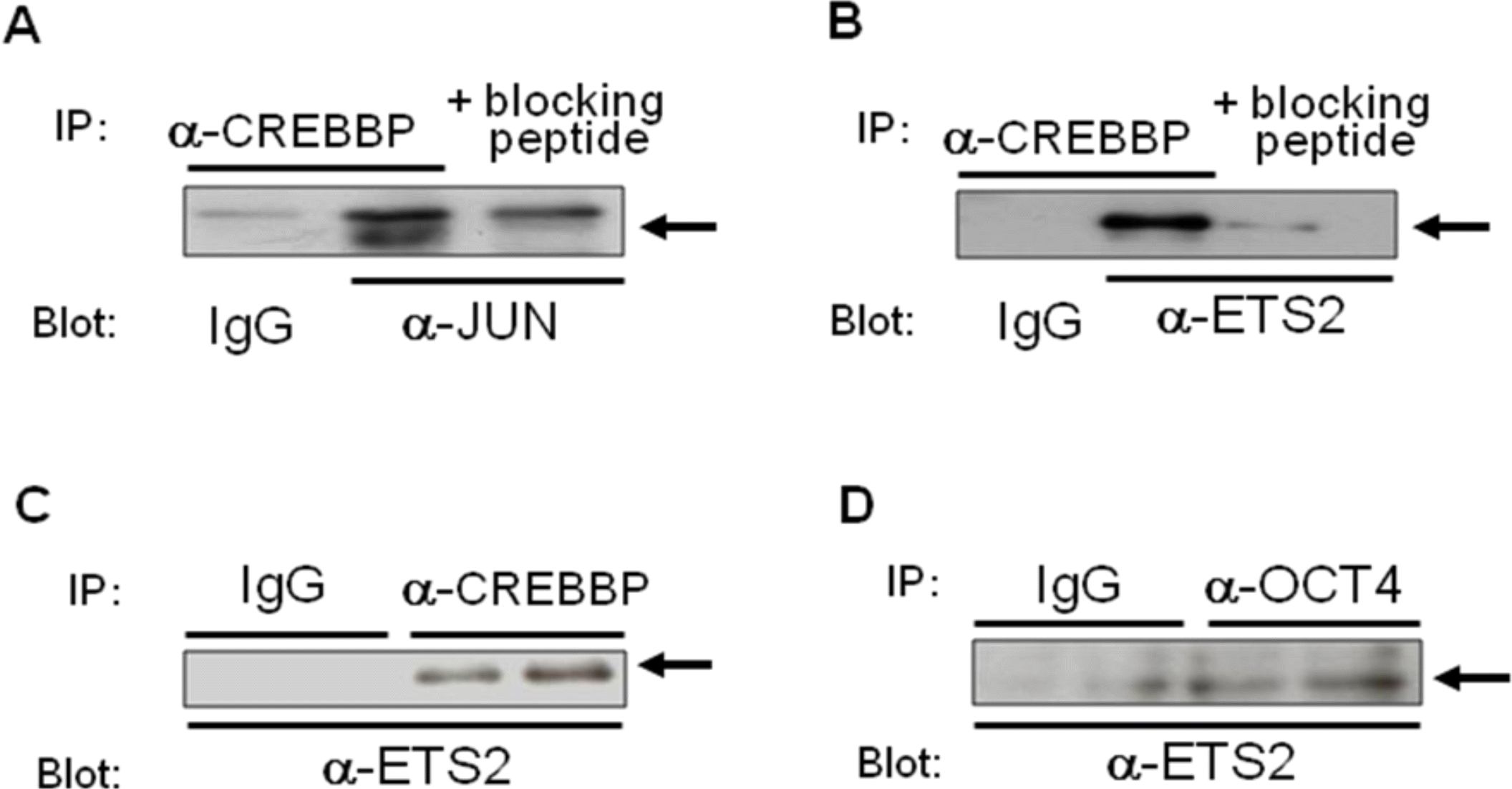
To determine whether either of these transcription factors directly interact with CREBBP, thereby resulting in changes in ovine IFNT gene transcription, co-immunoprecipitation studies were carried out. Targeted transcription factors were co-transfected into JEG-3 cells, of which optimum expression was predetermined for each expression plasmid. The co-immunoprecipitation study (Figure 6) showed that JUN and ETS2 bound to CREBBP, and binding between OCT4 and ETS2 was also demonstrated.
It was previously thought that OCT4 expression subsides long before the elongation of bovine conceptuses begins (Ezashi et al., 2001). However, recent studies have shown that OCT4 expression persists much longer in ruminant conceptuses (Degrelle et al., 2005; Berg et al., 2011). This study represents the first to examine direct interactions between OCT4, CDX2, and CREBBP within the context of IFNT transcriptional regulation. Our data showed that cotransfection of Cdx2 with Jun and Ets2 into JEG-3 cells resulted in the highest ovine IFNT gene transcription, which is in agreement with our previous study (Imakawa et al., 2006). Importantly, OCT4 exhibited inhibitory effects on ovine IFNT gene transcription only when Oct4 was transfected into JEG-3 cells prior to the other expression plasmids. These data imply that OCT4 binding to ETS2 (Ezashi et al., 2001) reduces the availability of ETS2 to form the transcription factor complex, JUN-CREBBP-ETS2, which is required for active IFNT transcription (Xu et al., 2003; Imakawa et al., 2006). Since the DNA-binding domain of ETS2 is located close to the region, to which OCT4 may bind (Ezashi et al., 2001), the OCT4-ETS2 complex may not be able to bind to the upstream region of the ovine IFNT gene. As CDX2 expression increases, CDX2 binds to residual OCT4 (Niwa et al., 2005), allowing binding of ETS2 to the JUN-CREBBP-ETS2 complex and activation of ovine IFNT gene transcription.
The importance of JUN-CREBBP-ETS2 complex formation for ovine IFNT gene transcription (Xu et al., 2003) was further evaluated by using a CREBBP inhibitor, 12S E1A. The binding of 12S E1A to CREBBP prevents CREBBP from forming JUN-CREBBP and/or ETS2-CREBBP complexes (Bannister and Kouzarides, 1995). In this study, cotransfection of Crebbp with Jun, Ets2 and Cdx2 did not further increase the level of ovine IFNT gene transcription. However, the transfection of 12S E1A reduced the level of ovine IFNT gene transcription by a greater extent than did Oct4 transfection. These observations support our hypothesis that disruption of the JUN-CREBBP-ETS2 complex formation results in inactivation of ovine IFNT gene transcription.
The presence of OCT4 mRNA in bovine and ovine trophoblasts detected in the present study and by others (Kurosaka et al., 2004; Degrelle et al., 2005; Berg et al., 2011; Saadeldin et al., 2012) contrasts with data from mouse trophoblasts, in which OCT4 expression is restricted to the ICM (Niwa et al., 2005). Berg et al. (2011) showed that the upstream conserved region 4 (CR4) of OCT4 genes differs between bovine and murine species, and that bovine CR4 is effective for OCT4 expression in mouse TE. Unlike murine blastocysts, ruminant conceptuses go through extensive trophoblast elongation prior to conceptus attachment to the uterine endometrium (Nagaoka et al., 2003). It has been suggested that there may be 2 types of trophoblast cell populations in the elongating blastocyst: terminally differentiated and less differentiated. We suspect that the terminally differentiated trophoblast cells produce IFNT, whereas less differentiated cell populations proliferate during the pre-attachment period.
CDX2 functions as a transcription factor that is required for IFNT transcription (Imakawa et al., 2006; Sakurai et al., 2009). In addition, CDX2 maintains the euchromatin state for IFNT transcription (Sakurai et al., 2010). CDX2 expression declines as conceptus attachment to the uterine epithelium proceeds. Thus, it is possible that reduced expression of CDX2 allows ETS2 to become available for JUN-CREBBP-ETS2 complex formation, while the euchromatin state maintained by CDX2 allows the complex to have full access to the upstream region of IFNT gene. Transcription factors and epigenetic regulation allow the TE to produce substantial amounts of IFNT during the conceptus elongation period.
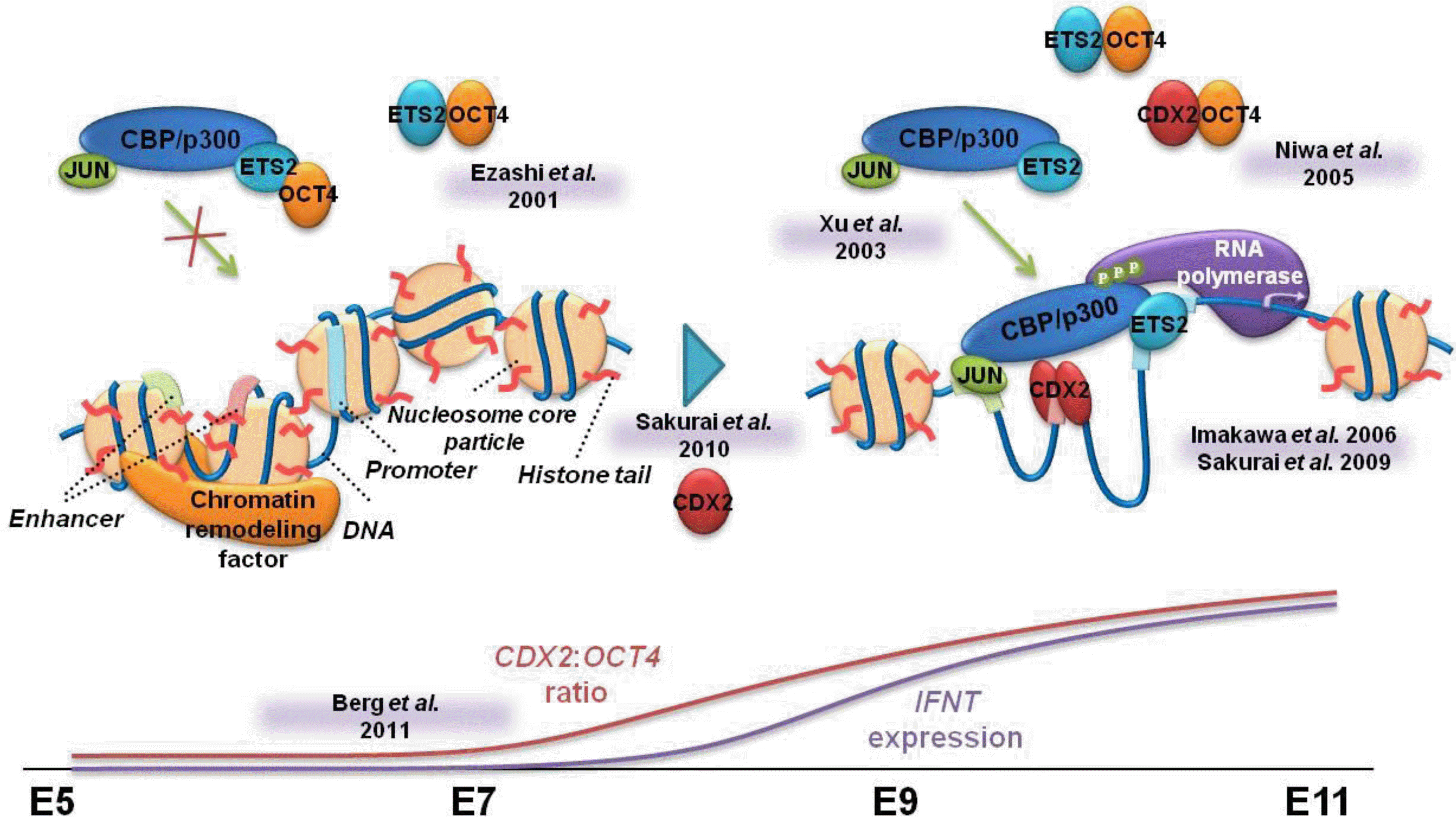
Based on these observations, we propose the following molecular mechanism for ovine IFNT gene transcription (Figure 7). When OCT4 expression exceeds that of CDX2 expression, OCT4 binds to ETS2, resulting in minimal formation of the transcription factor complex, JUN-CREBBP-ETS2. Increasing CDX2 expression, along with decreased levels of OCT4 protein, allows CDX2 to bind to residual OCT4, freeing up more ETS2 for formation of the JUN-CREBBP-ETS2 complex. This complex formation and the appropriate binding of these and other transcription factors to the upstream region of the ovine IFNT gene results in a high level of ovine IFNT gene transcription during the pre-attachment period, when the region is maintained in the euchromatin state.
ACKNOWLEDGMENTS
The initial portion of this study was supported in part by a Grant-in-Aid for Scientific Research (18108004 to KI) from the Japan Society for the Promotion of Science.
Figure┬Ā1.
Semi-quantitative RT-PCR showing the presence of transcription factor mRNAs in the ovine conceptuses and western blots showing proteins produced from expression vectors. CDX2 mRNA was found on days 15 and 17, while OCT4 mRNA was detected on days 15, 17, and 21 of gestation. During the same period, levels of CREBBP, JUN and ETS2 in ovine conceptuses were similar whereas IFNT mRNA declined (Figure 1A). The efficacy of all pSG5-based expression vectors was examined via transient transfection into JEG-3 cells, from which nuclear proteins (cell lysates) were isolated and subjected to western blot analysis. All expression constructs exhibited proteins of the expected sizes (Figure 1B). In both cases, one of four replicates is shown.
Figure┬Ā2.
The effect of transfection of Jun, Cdx2 and Ets2 along with Oct4 on IFNT gene transcription. The luciferase activity of -654-IFNT-Luc transfected with Jun, Ets2 and Cx2 expression plasmids was approximately 12 times that of the -654-IFNT-Luc plasmid alone. OCT4 inhibited -654-IFNT-Luc transcription when either Jun, Ets2, Cdx2, or Jun/Ets2, together with the reporter construct, was transfected into JEG-3 cells. However, OCT4 did not suppress the level of -654-IFNT-Luc transcription when Cdx2 was cotransfected with Ets2, Jun or Ets2/Jun into JEG-3 cells.

Figure┬Ā3.
Examination of the effect of JUN, CDX2, and/or ETS2 binding sites on IFNT gene transcription. Cotransfection of the transcription factor binding site-mutated -654-IFNT-Luc construct with Jun, Ets2 and Cdx2 reduced luciferase activities. However, cotransfection of the Oct4 expression plasmid along with Jun, Ets2 and Cdx2 did not change the degree of luciferase activity, which had already been reduced due to mutation of the transcription factor binding sites.
Figure┬Ā4.
Effects of various combinations of transcription factors that were transfected into JEG-3 cells at 0 or 24 h after initiation of the experiment. The maximum inhibitory effect of OCT4 on ovine IFNT gene transcription was found when Oct4 alone was transfected prior to any other transcription factor expression plasmids. From the 0/24 h experiments, the initial expression and/or complex formation between the transcription factors appeared to determine the subsequent degree of ovine IFNT gene transcription.
Figure┬Ā5.
Effect of CREBBP on ovine IFNT gene transcription. No additional increase in ovine IFNT gene transcription was observed when Crebbp was cotransfected with Jun, Ets2 and/or Cdx2. However, cotransfection of 12S E1A reduced the level of ovine IFNT gene transcription even in the presence of CDX2, and the level of luciferase activity reduction appeared to be more than that produced by Oct4 cotransfection.
Figure┬Ā6.
Determination of the relationship between JUN/ETS2 and/or CREBBP using co-immunoprecipitation. A. Jun and Crebbp expression plasmids were transfected into JEG-3 cells, from which the nuclear proteins were extracted. These extracts were subjected to immunoprecipitation with anti-CREBBP antibody, followed by western blot analysis with anti-JUN antibody. B. Ets2 and Crebbp expression constructs were transfected into JEG-3 cells, from which nuclear proteins were subjected to immunoprecipitation with anti-CREBBP antibody, followed by western blot analysis with anti-ETS2 antibody. C. Immunoprecipitation and western blot analyses were performed using nuclear extracts isolated from JEG-3 cells, into which Jun, Ets2, Cdx2, Crebbp, and Oct4 expression constructs were transfected. D. Ets2 and Oct4 expression plasmids were transfected into JEG-3 cells, from which nuclear proteins were subjected to immunoprecipitation with anti-OCT4 antibody, followed by western blot analysis with anti-ETS2 antibody. Binding between OCT4 and ETS2 was also demonstrated.
Figure┬Ā7.
Model showing the molecular mechanism of activation of ovine IFNT gene transcription. When OCT4 expression predominates in the ovine blastocyst, OCT4 binding to ETS2 (Ezashi et al., 2001) may limit the formation of the JUN-CREBBP-ETS2 complex (Xu et al., 2003). As CDX2 expression increases and OCT4 expression subsides, the CDX2 to OCT4 ratio increases (Berg et al., 2011), which loosens the chromatin structure (euchromatin state; Sakurai et al., 2010). These CDX2 proteins bind to residual OCT4 (Niwa et al., 2005), allowing ETS2 to form the JUN-CREBBP-ETS2 complex. The appropriate assembly of transcription factors at the euchromatin state results in maximum transcription of the ovine IFNT gene during the period of conceptus elongation.
Table┬Ā1.
Oligonucleotide primer sequences used for RT-PCR
REFERENCES
Bannister AJ, Kouzarides T. 1995. CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J 14:4758ŌĆō4762.



Beck F, Erler T, Russell A, James R. 1995. Expression of Cdx-2 in the mouse embryo and placenta: possible role in patterning the extra-embryonic membranes. Dev Dyn 204:219ŌĆō227.


Berg DK, Smith CS, Pearton DJ, Wells DN, Broadhurst R, Donnison M, Pfeffer PL. 2011. Trophectoderm lienage determination in cattle. Dev. Cell 20:244ŌĆō255.


Chawengsaksophak K, James R, Hammond VE, Kontgen F, Beck F. 1997. Homeosis and intestinal tumours in Cdx2 mutant mice. Nature 386:84ŌĆō87.


Degrelle SA, Campion E, Cabau C, Piumi F, Reinaud P, Richard C, Renard JP, Hue I. 2005. Molecular evidence for critical period in mural trophoblast development in bovine blastocysts. Dev Biol 288:448ŌĆō460.


Demmers KJ, Derecka K, Flint A. 2001. Trophoblast interferon and pregnancy. Reproduction 121:41ŌĆō49.


Ezashi T, Ealy AD, Ostrowski MC, Roberts RM. 1998. Control of interferon- gene expression by Ets2. Proc. Natl. Acad. Sci. USA 95:7882ŌĆō7887.



Ezashi T, Ghosh D, Roberts RM. 2001. Repression of Ets-2 induced transactivation of the interferon-promoter by Oct-4. Mol Cell Biol 21:7883ŌĆō7891.



Farin CE, Imakawa K, Roberts RM. 1989. In situ localization of mRNA for the interferon, ovine trophoblast protein-1, during early embryonic development of the sheep. Mol Endocrinol 3:1099ŌĆō1107.


Godkin JD, Bazer FW, Moffatt J, Sessions F, Roberts RM. 1982. Purification and properties of a major, low molecular weight protein relaeased by the trophoblast of sheep blastocysts at day 13ŌĆō21. J Reprod Fertil 65:141ŌĆō150.


Guillomot M, Michel C, Gaye P, Charlier N, Trojan J, Martal J. 1990. Cellular localization of an embryonic interferon, ovine trophoblastin and its mRNA in sheep embryos during early pregnancy. Biol. Cell 68:205ŌĆō211.


Imakawa K, Anthony RV, Kazemi M, Marotti KR, Polites HG, Roberts RM. 1987. Interferon-like sequence of ovine trophoblast protein secreted by embryonic trophectoderm. Nature 330:377ŌĆō379.


Imakawa K, Kim MS, Matsuda-Minehata F, Ishida S, Iizuka M, Suzuki M, Chang KT, Echternkamp SE, Christenson RK. 2006. Regulation of the ovine interferon-tau gene by a blastocyst-specific transcription factor, Cdx2. Mol Reprod Dev 73:559ŌĆō567.


Kurosaka S, Eckardt S, McLaughlin KJ. 2004. Pluriopotent lineage definition in bovine embryos by Oct4 transcript localization. Biol Reprod 71:1578ŌĆō1582.


Matsuda F, Xu N, Kijima S, Tachi C, Christenson RK, Sakai S, Imakawa K. 2004. Analysis of transcriptional control elements in the 5ŌĆÖ-upstream region of ovine interferon-gene using feeder-independent caprine trophoblast cell line, HTS-1. Placenta 25:166ŌĆō175.


Nagaoka K, Nojima H, Watanabe F, Chang KT, Christenson RK, Sakai S, Imakawa K. 2003. Regulation of blastocyst migration, apposition, and initial adhesion by a chemokine, interferon-inducible protein 10 kDa (IP-10), during early gestation. J Biol Chem 278:29048ŌĆō29056.


Nephew KP, Whaley AE, Christenson RK, Imakawa K. 1993. Differential expression of distinct mRNAs for ovine trophoblast protein-1 and related sheep type I interferons. Biol Reprod 48:768ŌĆō778.


Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95:379ŌĆō391.


Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. 2005. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123:917ŌĆō929.


Palmieri SL, Peter W, Hess H, Scholer HR. 1994. Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev Biol 166:259ŌĆō267.


Roberts RM, Cross JC, Leaman DW. 1992. Interferons as hormones of pregnancy. Endocr Rev 13:432ŌĆō452.


Ralston A, Rossant J. 2005. Genetic regulation of stem cell origins in the mouse embryo. Clin Genet 68:106ŌĆō112.


Saadeldin IM, Choi W, da Torre BR, Kim B, Lee B, Jang G. 2012. Embryonic development and implantation related gene expression of oocyte reconstructed with bovine trophoblast cells. J Reprod Dev 58:425ŌĆō431.


Sakurai T, Sakamoto A, Muroi Y, Bai H, Nagaoka K, Tamura K, Takahashi T, Hashizume K, Sakatani M, Takahashi M, Godkin JD, Imakawa K. 2009. Induction of endogenous interferon tau gene transcription by CDX2 and high acetylation in bovine nontrophoblast cells. Biol Reprod 80:1223ŌĆō1231.


Sakurai T, Bai H, Konno T, Ideta A, Aoyagi Y, Godkin JD, Imakawa K. 2010. Function of a transcription factor CDX2 beyond its trophectodermal lienage specification. Endocrinology 151:5873ŌĆō5881.


Stewart HJ, McCann SH, Barker PJ, Lee KE, Lamming GE, Flint AP. 1987. Interferon sequence homology and receptor binding activity of ovine trophoblast antiluteolytic protein. J Endocrinol 115:R13ŌĆōR15.


Suh E, Chen L, Taylor J, Traber PG. 1994. A homeodomain protein related to caudal regulates intestine-specific gene transcription. Mol Cell Biol 14:7340ŌĆō7351.



Xu N, Takahashi Y, Matsuda F, Sakai S, Christenson RK, Imakawa K. 2003. Coactivator CBP in the regulation of conceptus IFN gene transcription. Mol Reprod Dev 65:23ŌĆō29.


- TOOLS





 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print





