 |
 |
| Anim Biosci > Volume 36(10); 2023 > Article |
|
Abstract
Objective
The study investigated the origin of the Akhal-Teke horse using genome-wide single-nucleotide polymorphism (SNP) data and mitochondrial hypervariable region 1 (HVR-1) nucleotide sequences
Methods
Genome-wide SNP data from 22 breeds (481 horses) and mitochondrial HVR-1 sequences from 24 breeds (544 sequences) worldwide to examine the origin of the Akhal-Teke horse. The data were analyzed using principal component analysis, linkage disequilibrium analysis, neighbor-joining dendrograms, and ancestry inference to determine the population relationships, ancestral source, genetic structure, and relationships with other varieties.
Results
A close genetic relationship between the Akhal-Teke horse and horses from the Middle East was found. Analysis of mitochondrial HVR-1 sequences showed that there were no shared haplotypes between the Akhal-Teke and Tarpan horses, and the mitochondrial data indicated that the Akhal-Teke horse has not historically expanded its group. Ancestral inference suggested that Arabian and Caspian horses were the likely ancestors of the Akhal-Teke horse.
The Akhal-Teke horse is estimated to be at least 3,000 years old and is one of the oldest horse breeds in the world. Native to the southern region of Turkmenistan, the Akhal-Teke horse is adapted to harsh climatic conditions [1] and is famous for its beautiful appearance, familiarity with people, and shining coat color. The Akhal-Teke horse is one of the finest horse breeds in the world. Today, the Akhal-Teke horse population is slowly declining and is endangered. Currently, the Akhal-Teke is found mainly in Turkmenistan [2].
The Akhal-Teke is the best of a number of excellent horse breeds. The origin of this horse breed is highly controversial. Several hypotheses regarding the origin of the Akhal-Teke horse have been proposed based on archaeological and documentary evidence. The most plausible hypothesis is that the Akhal-Teke horse originated in Central Asia, in the area between the Caspian coast and the Fergana region [3]. It is thought that the earliest domestication of horses occurred in the Eurasian steppe around 3500 BCE, from where they spread across the Eurasian continent [4]. A study of DNA microsatellite genotyping of 2,024 Akhal-Teke horses from around the world showed that this horse breed has a high level of genetic diversity with abundant rare alleles at microsatellite DNA loci. Further analysis of the rare alleles showed that the Akhal-Teke horse is very similar to Tuvinia and Khakassia horses [5]. These results support the hypothesis of a Central Asian origin; studies investigating population changes associated with domestication from 273 ancient horse genomes also suggest that the western Eurasian steppe region is the home of the Akhal-Teke horse [6]. In contrast, there is a great deal of evidence indicating that the Akhal-Teke horse originated in the Middle East. The horse was widely documented in Middle Eastern epics and was referred to as the “supernatural horse,” “celestial horse,” or “bleeding stallion” [7]. Various molecular markers have been used in previous studies, which suggested that the Akhal-Teke horse belongs to the Middle Eastern horse breed clade [8–11]. Other archaeological studies at various sites suggest that the Akhal-Teke horse originated in Southwest Asia and migrated from Central Asia to the Middle East, or originated as a cross between the Tarpan and a domesticated native horse from Central Asia [12]. Thus, although several hypotheses have been proposed for the origin of the Akhal-Teke horse, many aspects surrounding its origin are unknown. In this study, we combined genotypic data from genome-wide high-density single-nucleotide polymorphism (SNP) arrays with equine mitochondrial hypervariable regions 1 (HVR-1) sequence data to elucidate the origin of the Akhal-Teke. The results provide new insights into the origin of the Akhal-Teke horse.
The experiment was approved by the Institutional Animal Care and Use Committee of the Gansu Agricultural University under permit number No.GAU-LC-2018-12.
The genome-wide SNP genotype data for Kazakh and Przewalskii horses were obtained from our previous study [13] that used the Illumina horse SNP 70 bead array (Illumina, San Diego, CA, USA) with the Illumina Equine SNP 50 bead array data (animalgenome.org) (Illumina, USA) from 20 Western horse breeds genotyped by Petersen et al [14,15]. The SNP70 and SNP50 arrays shared a total of 43,782 common markers and were used in subsequent analyses (Table 1). Quality control of the raw data was performed with PLINK v1.07 [16] and SNPs with call rates lower than 0.90, p<10−5 or MAF<0.01 were excluded. Finally, 36,734 sites remained after quality control.
PopLDdecay software was used to measure the R2 of each population [17]. A curve of linkage disequilibrium (LD) change was drawn with R2 as the vertical axis and the physical distance between paired SNPs (<400 kb) as the horizontal axis.
The frequency of shared haplotypes between Akhal-Teke horses and other horse breeds was calculated using IBDLD software (v3.37) [18]. The specific parameters were set as follows: “plinkbf int evolution -method GIBDLD -ploci 10 -nthreads 24 -step 0 -hiddenstates 3 -segment -length 10 -min 0.8”. The normalized identity by descent (nIBD) parameter was used for the statistics of haplotypes shared between Akhal-Teke horses and other horse breeds. The parameter Nibd = cIBD/tIBD was used in the formula, where cIBD represents all haplotypes shared by Akhal-Teke horses and other specific horse breeds, and tIBD is the number of pairs of Akhal-Teke and other specific horse breeds [19].
The F4 ratio in the ADMIXTOOLS software package [20] was used to predict the most likely ancestral population. Default parameters were used in the analysis: the ratio f4 (A, O; X, C)/f4 (A, O; B, C) was calculated, where population X is the Akhal-Teke horse, A is the Exmoor horse with a relatively distant genetic relationship to X, O is the outgroup, the Przewalski horse, C is the thoroughbred, B is the other horse population The following is a list of the groups.
ALDER v1.03 [21] software was used to estimate the underlying proportions of genetic contributions from other horse breeds to the Akhal-Teke horse. This software determines the optimal ancestry in a reference population based on the difference in allele frequencies in the ancestral population and the correlation of SNPs in a weighted mixed target population. Here, Arabian and Caspian horses were used as the reference populations, and the Akhal-Teke horse as the target population. The parameters were mincount = 4, binsize = 0.0005, maxdis = 0.5, and fast_snpread = NO.
The mitochondrial HVR-1 region sequences of 24 horse breeds from around the world were retrieved from the NCBI and manually edited and verified using DNASTAR software. Multiple alignments were performed using Mega7.0 to remove redundant sequences. The 24 horse breeds were divided into seven groups, namely, the Middle Eastern, Central Asian, European, Tarpan, Przewalski, and Akhal-Teke horse groups. The phylogenetic relationships were reconstructed using Popart software [22] and the differentiation index (FST) between populations was calculated using Arlquine software, while mismatch analysis was performed for the Akhal-Teke horse to detect if there were expansion events in the history of the horse population.
The results of the nearest neighbor-joining tree showed that Akhal-Teke horses and Arabian horses form a cluster together, indicating that they are closely related. However, no close kinship was found between the Akhal-Teke horse and Caspian or Tuwa horses (Figure 1A), and principal component analysis results further confirmed the close genetic link between the Akhal-Teke horse and Middle Eastern horse populations (Figure 1B). Estimation of the genetic composition of the Akhal-Teke horse and other horses showed a minimum cross-validation statistic when K was set to 15. The results showed that the Akhal-Teke horse had a similar genetic background to Middle Eastern horse breeds, especially Arabian horses (Figure 1D). The Treemix results reflect several well-known gene flows (Figure 2A). For example, Thoroughbreds were observed to have had a significant genetic influence on Quarter horses, and Caspian horses were influenced by gene flow from Arabian horses. However, no gene exchange between Akhal-Teke and other horse breeds was detected and LD decay analysis showed that Akhal-Teke horses had LD decay levels similar to those of Tuva, Caspian, and Arabian horses. The results of this analysis indicated a moderate level of LD decay in the Akhal-Teke horses(Figure 2B).
The results of the IBD shared-fragment analysis indicated the presence of relatively large shared fragments between the Akhal-Teke horse and Arabian horses and their derivatives (Thoroughbreds and Quarter horses). On the other hand, the length of shared fragments between the Tuva and Caspian horses and the Akhal-Teke horses was small. Nordic horses, especially the Exmoor breed, shared the fewest fragments with the Akhal-Teke horse (Figure 3).
While the above analysis indicated a close genetic relationship between the Akhal-Teke horse and the Arabian horse and several other breeds, its ancestry was still undetermined. Therefore, the ADMIXTOOLS software package was applied to analyze the best ancestral composition. Exmoor and Thoroughbred horses were used as reference populations and the Akhal-Teke horse was used as the target population (X). The higher the output f4 ratio, the greater the likelihood of the reference population being ancestral to the target population. The results showed that Middle Eastern horses had a larger f4 ratio than the Central Asian horse population (Table 2).
The horse groups most likely to be ancestral to the Akhal-Teke horse were selected from the Middle Eastern and Central Asian horse populations. Alder v103 software was used to estimate the ancestral ratios of Arabian horses and Caspian horses in the Akhal-Teke horse. The Caspian and Arabian horses were used as the two reference groups, and the Akhal-Teke horse was used as the target group. The results showed that Arabian horses accounted for 26% of the ancestry of Akhal-Teke horses and Caspian horses accounted for 18% of the ancestry of Akhal-Teke horses (Figure 4).
The 544 hypervariable regions of the 24 breeds were classified into seven major groups according to the geographical location of the population. These groups were the Middle Eastern, Akhal-Teke, Central Asian, European, Tarpan, Przewalski, and Thoroughbred horse groups. A total of 175 haplotypes were detected (Figure 5A). The results of the central join network showed that Akhal-Teke horses shared several haplotypes with horses from the Middle East and Central Asia. However, there were no common haplotypes between Akhal-Teke horses and Tarpan horses. The 175 haplotypes were also divided into five haplotype groups, A–E, with Tarpan horses found only in the E haplotype group and other horses marked for inclusion. The results of genetic differentiation between groups (FST) analysis showed that the FST values between the Akhal-Teke and Tarpan, Przewalski, and Andalusian horses were relatively high compared to the other horse populations (Figure 5B). Mismatch analysis of the Akhal-Teke horse population showed multiple peaks (Figure 5C), indicating that there was no expansion in the history of this breed. Ultimately, the hypotheses that the Akhal-Teke horse is a cross between a local Central Asian breed and the Tarpan horse or that the Akhal-Teke horse migrated from Central Asia to the Middle East were not supported.
The Akhal-Teke horse is one of the oldest and most famous horse breeds. It is now mainly distributed in Turkmenistan [2]. Most of the horses have golden chestnut hair, sometimes black or brown. The shoulder height is 150 to 163 cm. Akhal-Teke horses belong to the riding-horse category due to their sporting characteristics [23]. However, despite being one of the few purebred horse breeds in the world, the origin of the Akhal-Teke horse is largely unknown. The most likely hypothesis is that the Akhal-Teke horse originated in Central Asia, adjacent to the Eurasian steppe, where the earliest domestication of horses took place [24]. Conversely, the hypothesis that the Akhal-Teke originated in the Middle East is less plausible, given the paucity of archaeological evidence associated with Akhal-Teke horses found in the Middle East [12]. As the Turkmen horse [25] is the most important ancestor of the modern Akhal-Teke horse [26], another proposal is that the Akhal-Teke horse originated in Central Asia and was introduced to the Middle East. To further test this hypothesis, this study applied genome-wide SNP data from 22 breeds from around the world to analyze the genetic relationship between the Akhal-Teke horse and other breeds. The results suggested that the Akhal-Teke horse is not closely related to the Caspian or Tuva horse and that there was little genetic exchange between the two, which does not support the longstanding hypothesis that the Akhal-Teke horse originated in Central Asia [27]. However, closer genetic links between the Akhal-Teke horse and horses from the Middle East were found. Genetic exchanges between the Akhal-Teke horse and other horse breeds could not be detected by the Treemix analysis, which may be due to the relatively limited geographic distribution of the Akhal-Teke horse. Shared IBD fragment analysis of the Akhal-Teke horse provided further evidence that Middle Eastern horses are more likely to be the ancestors of the Akhal-Teke horse than other horses with the admixture results indicating that both Caspian and Arabian horses have similar genetic backgrounds to the Akhal-Teke horse. Furthermore, the ancestry estimates showed that Arabian horses contributed a greater proportion to the ancestry of the Akhal-Teke horse. The geographical distributions of the Arabian and Akhal-Teke horses almost overlap, which would have facilitated genetic exchange between the two breeds. These results are consistent with fossil finds reported in the literature that the Libyan peninsula, adjacent to the Middle East, played an essential role in the process of horse domestication as a major center of domestication [24]. The fact that the modern Akhal-Teke breed shares haplogroups with horses from Turkmenistan and Kazakhstan is more clearly delineated by the mitochondrial data, which also points to the likelihood that the modern Middle East is the likely origin of the Akhal-Teke horse [2,10].
Due to the lack of whole-genome SNP array data for the Tarpan horse, data on the mitochondrial HVR-1 region of the Tarpan horse [16,28–30] was obtained from NCBI. The genetic relationship between the Akhal-Teke and Tarpan horses was examined. The haplotypes were divided into five groups. All Tarpan horses were found in the E haplotype group, and no haplotypes were shared between Tarpan horses and other horses [29]. Therefore, our mtDNA results do not support the hypothesis that the Akhal-Teke horse is a hybrid between the Tarpan horse and a local Central Asian horse. This result is also consistent with the results from the genome-wide SNP data analysis in this study. Furthermore, the mtDNA data also showed that there was no historical group expansion of the Akhal-Teke horse, which did not support the hypothesis that the Akhal-Teke migrated from Central Asia to the Middle East. Thus, the results from both the genomic and mtDNA data suggest that the Akhal-Teke most likely originated in the Middle East.
In this study, data from horse whole-genome SNP array chips and mitochondrial hypervariable regions indicated that the Akhal-Teke horse may have originated in the Middle East rather than in Central Asia. The study also found no support for the hypothesis that the breed was derived from a historical migration from Central Asia to the Middle East, or that it originated from a hybrid between the Tarpan horse and a domesticated local horse breed from Central Asia.
Notes
AUTHOR CONTRIBUTIONS
ZK, JS, and SC contributed to study design, collected biologic materials, clinical and demographic data, participated in writing the manuscript, and conceived and designed the experiments. QZ, YZ performed the experiments and analyzed the data. TL, JW, and ZL revised the paper. All authors have read and approved the final manuscript.
DATA AVAILABILITY STATEMENT
The access numbers of 544 mitochondrial HVR-1 sequences of 24 breads from NCBI are summarized in the excel table. Mitochondrial HVR-1 Nucleotide Sequence.xlsx
Figure 1
Genetic relationships between the Akhal-Teke horse and other horse breeds of the world. (A) Relationship between the Akhal-Teke horse and other horse populations shown by a neighbor-joining phylogenetic tree. Study populations are shown in different colors. (B) Principal component analysis of the Akhal-Teke horse and other horse populations. The different study populations are indicated by different colors. (C) Cross-validation under different K-values. (D) ADMIXTURE analysis of the Akhal-Teke horse and world horse populations. (K) Admixture plots of breeds analyzed with different numbers of assumed ancestors. Each column represents an individual. Columns represent individuals, groups of columns represent horse populations.
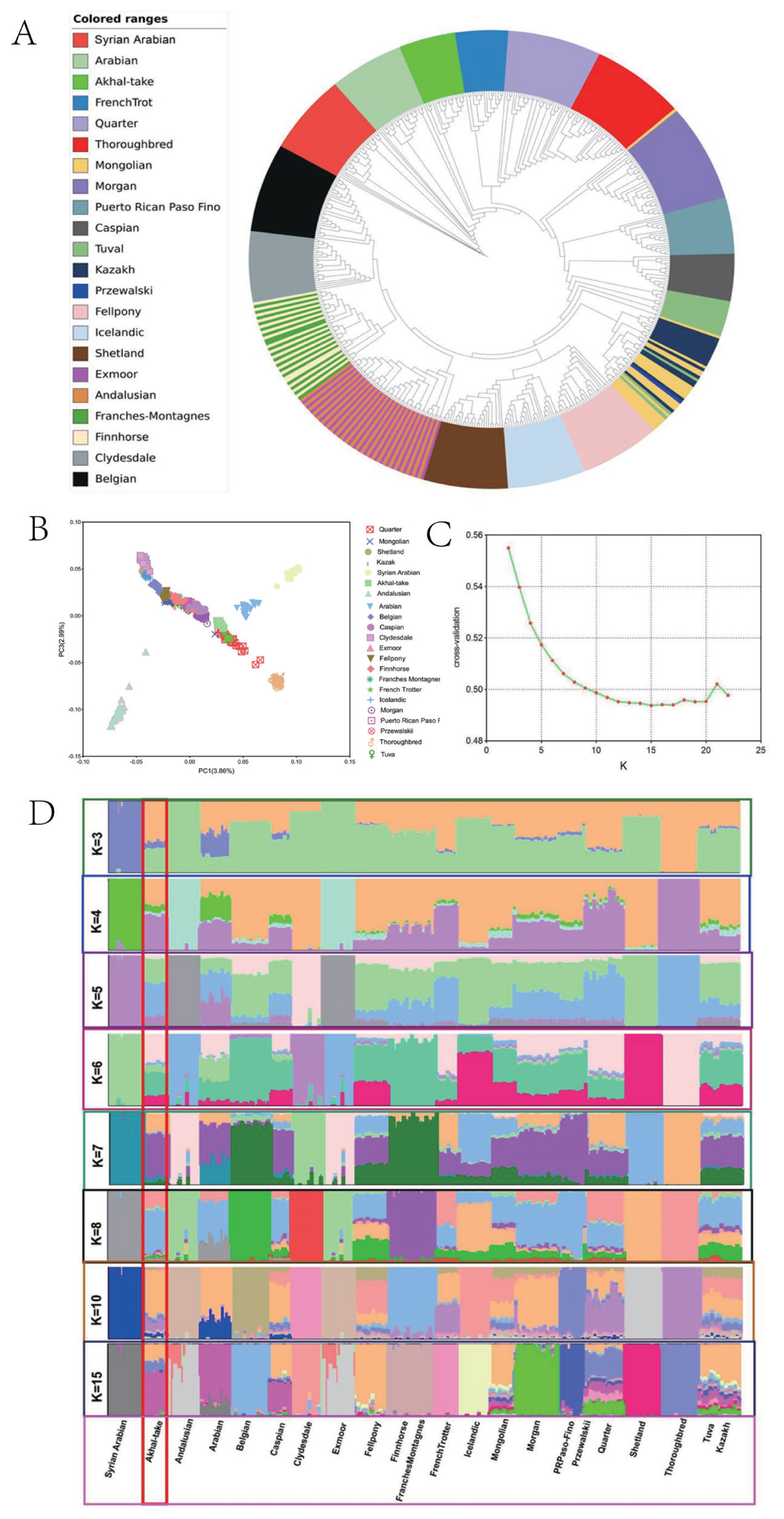
Figure 2
Population gene flow and the degree of domestication. (A) Migration showing four migration events in the Akhal-Teke horse and worldwide horse populations, shown by the TreeMix program; (B) linkage disequilibrium (LD) decay analysis of 22 horse populations from around the world, including Akhal-Teke horses. The different horse populations are represented by different colors.

Figure 3
Estimated lengths of identity by descent (IBD) segments common to the Akhal-Teke and other horse breeds from around the world.

Figure 4
Evaluation of ancestry ratios of the Akhal-Teke horse. Arabian and Caspian horses were used as the reference populations, and the Akhal-Teke horse as the target population. Ancestry ratios are indicated by a percent sign.
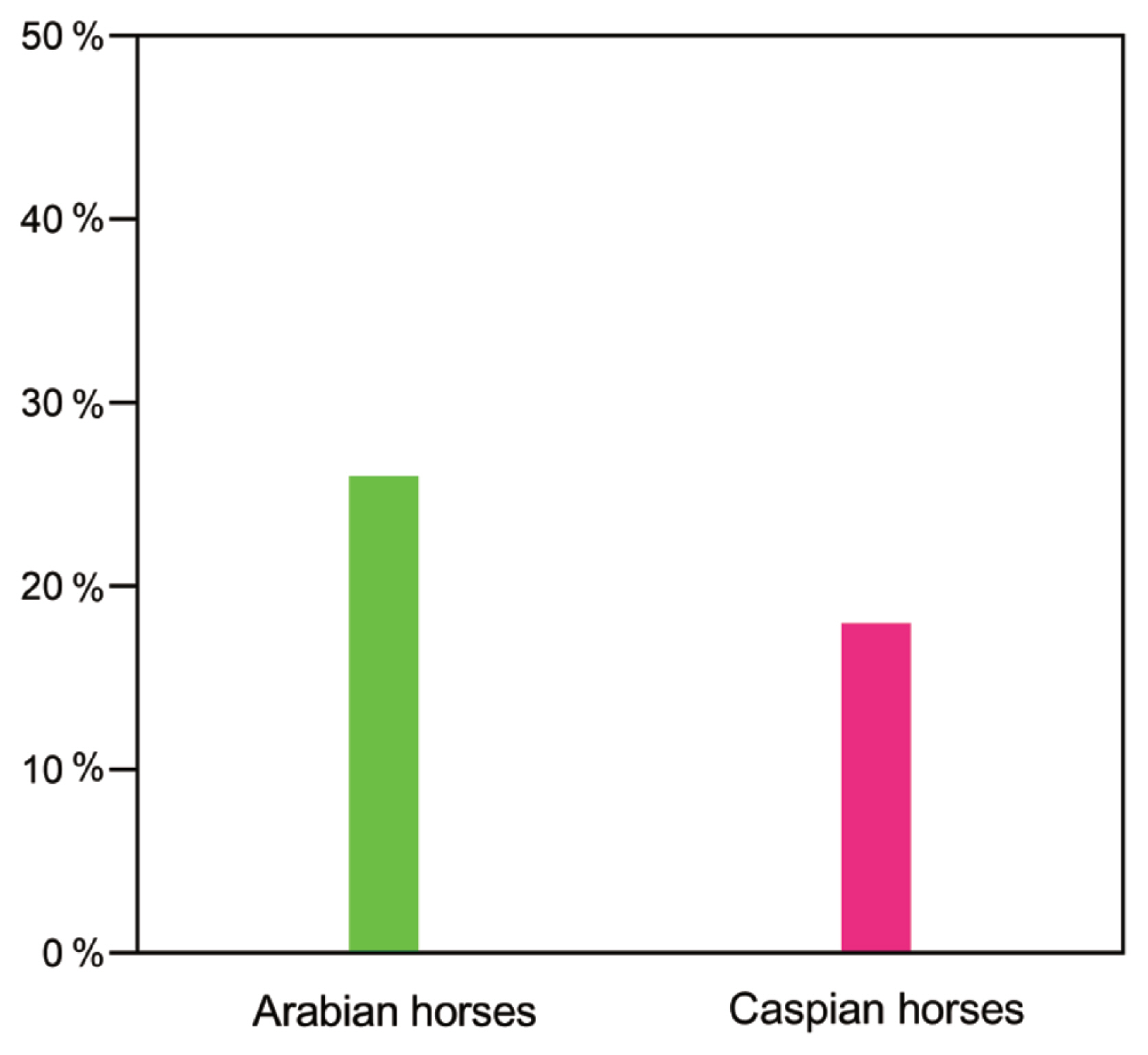
Figure 5
Results of the mitochondrial HVR-1 nucleotide sequence analysis. (A) Median-joining network between groups constructed in the mitochondrial HVR-1 region. Study populations are represented by different colors. All haplotypes were divided into five haplotype subgroups, with each subgroup circled and represented by a different letter. (B) Indicators of genetic differentiation between horse groups (FST). Color intensity indicates the degree of differentiation between groups. (C) Mismatch analysis diagram of the Akhal-Teke horse. HVR-1, hypervariable region 1.
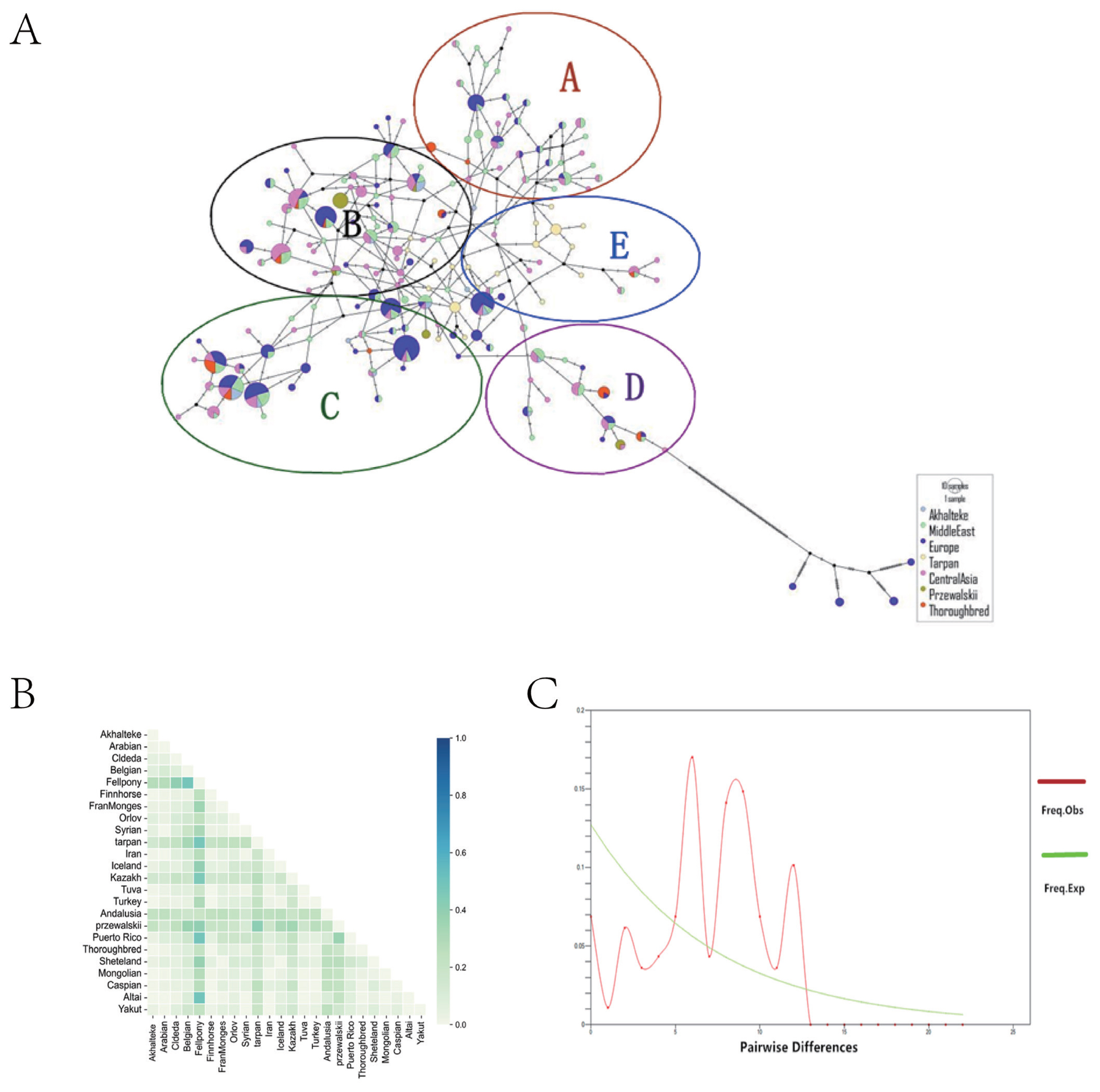
Table 1
Information on the horse breeds included in this study
| Breed | Population size | Geographic origin | Data source |
|---|---|---|---|
| Syrian Arabian | 27 | Syrian | Almarzook et al [15] |
| Akhal-Teke | 18 | Turkmenistan | Petersen et al [14] |
| Andalusian | 24 | Spain | Petersen et al [14] |
| Arabian | 24 | Middle East | Petersen et al [14] |
| Belgian | 30 | Belgium | Petersen et al [14] |
| caspian | 16 | Persia | Petersen et al [14] |
| Clydesdale | 24 | Scotland | Petersen et al [14] |
| Exmoor | 24 | England | Petersen et al [14] |
| Fellpony | 26 | England | Petersen et al [14] |
| Finnhorse | 18 | Finland | Petersen et al [14] |
| Franches-Montagnes | 18 | Switzerland | Petersen et al [14] |
| FrenchTrotter | 17 | France | Petersen et al [14] |
| Icelandic | 25 | Iceland | Petersen et al [14] |
| Mongolian | 19 | Mongolian | Petersen et al [14] |
| Morgan | 33 | United States | Petersen et al [14] |
| Puerto RicanPaso Fino | 19 | Puerto Rico | Petersen et al [14] |
| Przewalskii | 1 | China | Petersen et al [14] |
| Quarter | 30 | United States | Petersen et al [14] |
| Shetland | 27 | Scotland | Petersen et al [14] |
| Thoroughbred | 30 | United States | Petersen et al [14] |
| Tuva | 15 | Siberia | Petersen et al [14] |
| Kazakh | 16 | Middle East | Ma et al [13] |
Table 2
Analysis of the F4 ratio of Akhal-Teke horses
REFERENCES
1. Li B, He X, Zhao Y, et al. Analysis of the miRNA transcriptome during testicular development and spermatogenesis of the Mongolian horse. Reprod Fertil Dev 2020; 32:582–593.
https://doi.org/10.1071/RD19133


2. Cozzi MC, Strillacci MG, Valiati P, Rogliano E, Bagnato A, Longeri M. Genetic variability of Akhal-Teke horses bred in Italy. PeerJ 2018; 6:e4889
https://doi.org/10.7717/peerj.4889



3. Librado P, Gamba C, Gaunitz C, et al. Ancient genomic changes associated with domestication of the horse. Science 2017; 356:442–5.
https://doi.org/10.1126/science.aam5298


4. Cozzi MC, Strillacci MG, Valiati P, Bighignoli B, Cancedda M, Zanotti M. Mitochondrial D-loop sequence variation among Italian horse breeds. Genet Sel Evol 2004; 36:663
https://doi.org/10.1186/1297-9686-36-6-663



5. Leisson K, Alev K, Kaasik P, Jaakma Ü, Seene T. Myosin heavy chain pattern in the Akhal-Teke horses. Animal 2011; 5:658–62.
https://doi.org/10.1017/S1751731110002375


6. Cothran EG, Dyk E, Merwe FJ. Genetic variation in the feral horses of the Namib Desert, Namibia. J S Afr Vet Assoc 2001; 72:18–22.
https://doi.org/10.4102/jsava.v72i1.603


7. Achilli A, Olivieri A, Soares P, Torroni A. Mitochondrial genomes from modern horses reveal the major haplogroups that underwent domestication. Proc Natl Acad Sci USA 2012; 109:2449–54.
https://doi.org/10.1073/pnas.1111637109



8. Luís C, Juras R, Oom MM, Cothran EG. Genetic diversity and relationships of Portuguese and other horse breeds based on protein and microsatellite loci variation. Anim Genet 2007; 38:20–7.
https://doi.org/10.1111/j.1365-2052.2006.01545.x


9. Bömcke E, Gengler N, Cothran EG. Genetic variability in the Skyros pony and its relationship with other Greek and foreign horse breeds. Genet Mol Biol 2011; 34:68–76.
https://doi.org/10.1590/S1415-47572010005000113



10. Priskin K, Szabó K, Tömöry G, et al. Mitochondrial sequence variation in ancient horses from the Carpathian Basin and possible modern relatives. Genetica 2010; 138:211–8.
https://doi.org/10.1007/s10709-009-9411-x


11. Giulotto E, Raimondi E, Sullivan KF, et al. The unique DNA sequences underlying equine centromeres. Prog Mol Subcell Biol 2017; 56:337–354.
https://doi.org/10.1007/978-3-319-58592-5_14


12. Atsenova N, Palova N, Mehandjyiski I, Neov B, Radoslavov G, Hristov P. The sequence analysis of mitochondrial dna revealed some major centers of horse domestications: the archaeologist’s cut. J Equine Vet Sci 2022; 109:103830
https://doi.org/10.1016/j.jevs.2021.103830


13. Ma H, Wu Y, Xiang H, et al. Some maternal lineages of domestic horses may have origins in East Asia revealed with further evidence of mitochondrial genomes and HVR-1 sequences. PeerJ 2018; 6:e4896
https://doi.org/10.7717/peerj.4896



14. Petersen JL, Mickelson JR, Cothran EG, et al. Genetic diversity in the modern horse illustrated from genome-wide SNP data. PLoS One 2013; 8:e54997
https://doi.org/10.1371/journal.pone.0054997



15. Almarzook S, Reissmann M, Arends D, Brockmann GA. Genetic diversity of Syrian Arabian horses. Anim Genet 2017; 48:486–9.
https://doi.org/10.1111/age.12568


16. Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81:559–75.
https://doi.org/10.1086/519795



17. Zhang C, Dong SS, Xu JY, He WM, Yang TL. PopLDdecay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019; 35:1786–8.
https://doi.org/10.1093/bioinformatics/bty875


18. Ma H, Wang S, Zeng G, et al. The origin of a coastal indigenous horse breed in china revealed by genome-wide SNP data. Genes 2019; 10:241
https://doi.org/10.3390/genes10030241



19. Ablondi M, Dadousis C, Vasini M, Eriksson S, Mikko S, Sabbioni A. Genetic diversity and signatures of selection in a Native Italian horse breed based on SNP data. Animals (Basel) 2020; 10:1005
https://doi.org/10.3390/ani10061005



20. Petr M, Vernot B, Kelso J. admixr-R package for reproducible analyses using ADMIXTOOLS. Bioinformatics 2019; 35:3194–5.
https://doi.org/10.1093/bioinformatics/btz030



21. Loh PR, Lipson M, Patterson N, et al. Inferring admixture histories of human populations using linkage disequilibrium. Genetics 2013; 193:1233–54.
https://doi.org/10.1534/genetics.112.147330



22. Bertholet J, Anastasi G, Noble D, et al. Patterns of practice for adaptive and real-time radiation therapy (POP-ART RT) part II: Offline and online plan adaption for interfractional changes. Radiother Oncol 2020; 153:88–96.
https://doi.org/10.1016/j.radonc.2020.06.017



23. Lau AN, Peng L, Goto H, Chemnick L, Ryder OA, Makova KD. Horse domestication and conservation genetics of Przewalski’s horse inferred from sex chromosomal and autosomal sequences. Mol Biol Evol 2009; 26:199–208.
https://doi.org/10.1093/molbev/msn239


24. Warmuth V, Eriksson A, Bower MA, et al. Reconstructing the origin and spread of horse domestication in the Eurasian steppe. Proc Natl Acad Sci USA 2012; 109:8202–6.
https://doi.org/10.1073/pnas.1111122109



25. Rahimi-Mianji G, Nejati-Javaremi A, Farhadi A. Genetic diversity, parentage verification and genetic bottlenecks evaluation in Iranian Turkmen horse breed. Genetika 2015; 51:1066–74.
https://doi.org/10.7868/s0016675815090088


26. Orlando L. Ancient genomes reveal unexpected horse domestication and management dynamics. Bioessays 2020; 42:19400164
https://doi.org/10.1002/bies.201900164

27. Kusliy MA, Vorobieva NV, Tishkin AA, et al. Traces of late bronze and early iron age mongolian horse mitochondrial lineages in modern populations. Genes (Basel) 2021; 12:412
https://doi.org/10.3390/genes12030412



28. Yang L, Kong X, Yang S, et al. Haplotype diversity in mitochondrial DNA reveals the multiple origins of Tibetan horse. PLoS One 2018; 13:e0201564
https://doi.org/10.1371/journal.pone.0201564



29. Lippold S, Matzke NJ, Reissmann M, Hofreiter M. Whole mitochondrial genome sequencing of domestic horses reveals incorporation of extensive wild horse diversity during domestication. BMC Evol Biol 2011; 11:328
https://doi.org/10.1186/1471-2148-11-328



30. Csizmár N, Mihók S, Jávor A, Kusza S. Genetic analysis of the Hungarian draft horse population using partial mitochondrial DNA D-loop sequencing. PeerJ 2018; 6:e4198
https://doi.org/10.7717/peerj.4198



- TOOLS






 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print





