 |
 |
| Anim Biosci > Volume 36(8); 2023 > Article |
|
Abstract
Objective
Methods
Results
Conclusion
Notes
AUTHOR CONTRIBUTIONS
J.-N.O. and C.-K.L. conceptualized the study. J.-N.O. conducted overall experiments and data curation. M.L., G.C.C., D.-K.L., K.-H.C., S.-H.K., J.J. participated on writing-review and editing. D.-K.L., K.-H.C., and C.-K.L. supervised research. All authors have read and agreed to the published version of the manuscript.
CONFLICT OF INTEREST
We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.
FUNDING
This work was supported by the BK21 Four program, the Korea Evaluation Institute of Industrial Technology (KEIT) through the Alchemist project funded by the Ministry of Trade, Industry and Energy (MOTIE; 20012411), and the National Research Foundation of Korea (NRF) grant funded by the government of Republic of Korea (NRF-2021R1A2C 4001837).
SUPPLEMENTARY MATERIAL
Figure 1
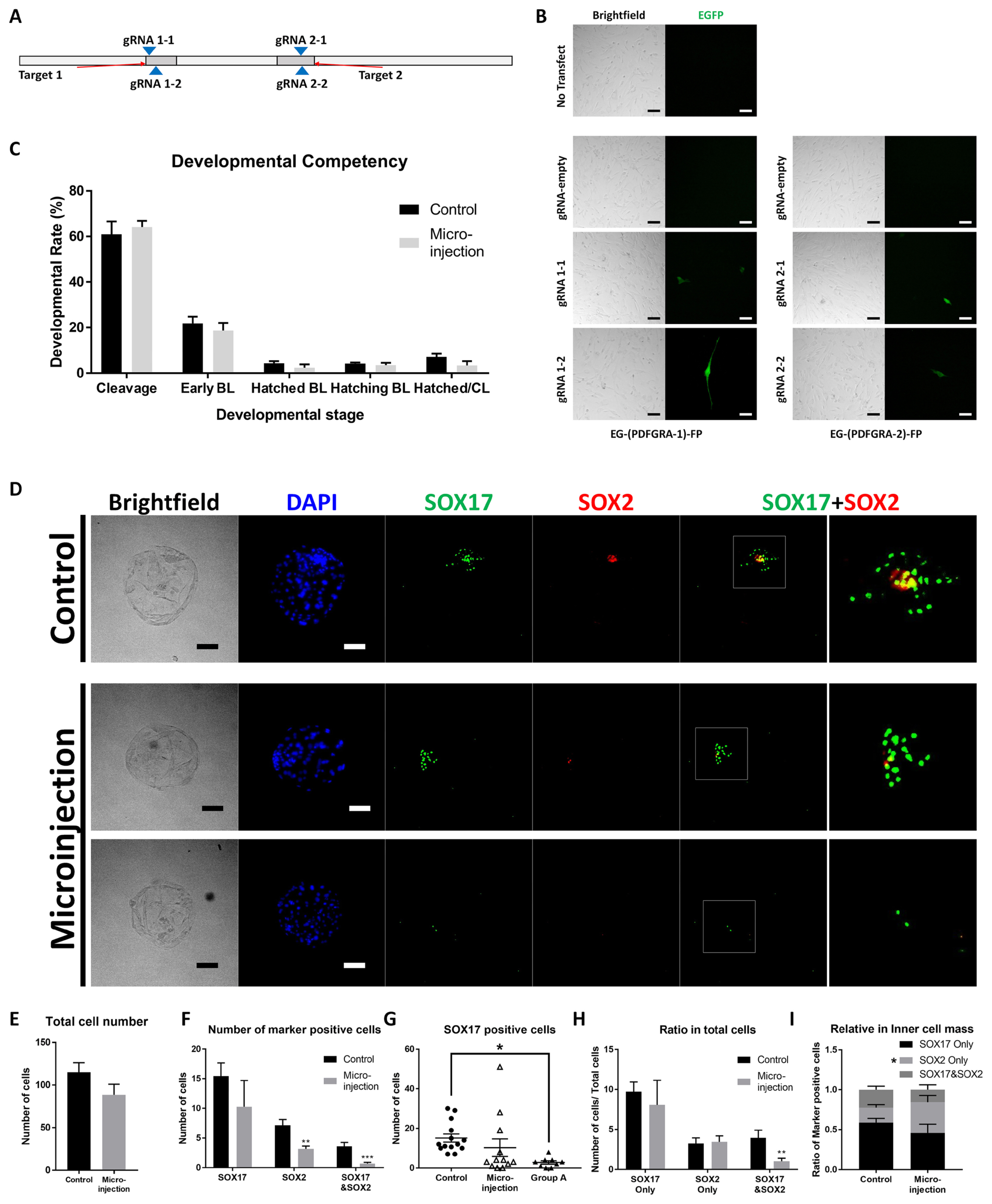
Figure 2
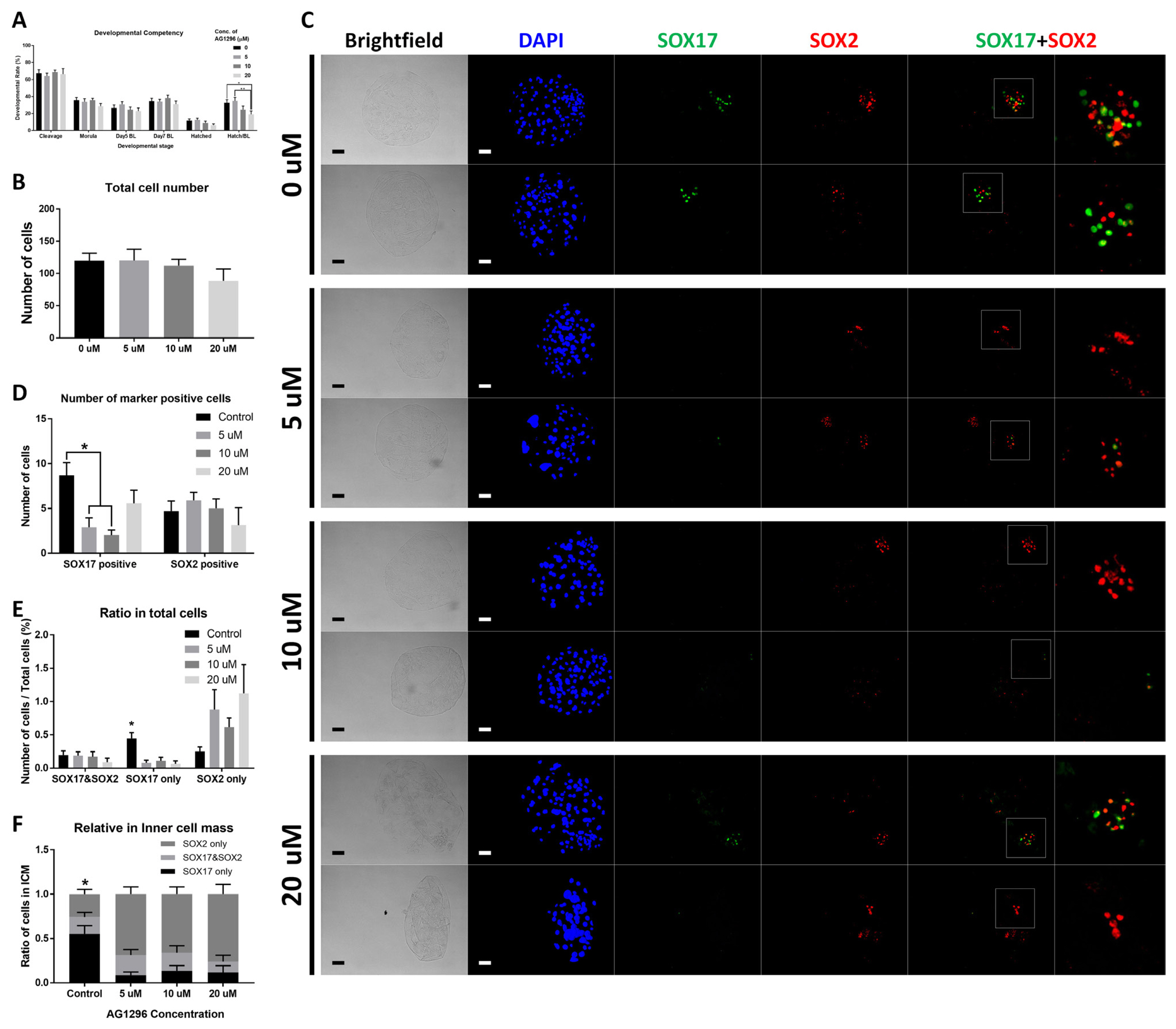
Figure 3

Table 1
Table 2
PCR, polymerase chain reaction; Gapdh, glyceraldehyde-3-phosphate dehydrogenase; Zfand5, zinc finger AN1-type containing 5; Myo1e, myosin IE; Sgpl1, sphingosine-1-phosphate lyase 1; Tiparp, TCDD inducible poly(ADP-ribose) polymerase; Csrnp1, cysteine and serine rich nuclear protein 1; Plekha1, pleckstrin homology domain containing A1; Txnip, thioredoxin interacting protein.
Table 3
REFERENCES
- TOOLS






 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print





