 |
 |
| Anim Biosci > Volume 36(1); 2023 > Article |
|
Abstract
Objective
In this study, metabolites that changed in the rumen fluid, urine and feces of dairy cows fed different feed ratios were investigated.
Methods
Eight Holstein cows were used in this study. Rumen fluid, urine, and feces were collected from the normal concentrate diet (NCD) (Italian ryegrass 80%: concentrate 20% in the total feed) and high concentrate diet (HCD) groups (20%: 80%) of dairy cows. Metabolite analysis was performed using proton nuclear magnetic resonance (NMR) identification, and statistical analysis was performed using Chenomx NMR software 8.4 and Metaboanalyst 4.0.
Results
The two groups of rumen fluid and urine samples were separated, and samples from the same group were aggregated together. On the other hand, the feces samples were not separated and showed similar tendencies between the two groups. In total, 160, 177, and 188 metabolites were identified in the rumen fluid, urine, and feces, respectively. The differential metabolites with low and high concentrations were 15 and 49, 14 and 16, and 2 and 2 in the rumen fluid, urine, and feces samples, in the NCD group.
Conclusion
As HCD is related to rumen microbial changes, research on different metabolites such as glucuronate, acetylsalicylate, histidine, and O-Acetylcarnitine, which are related to bacterial degradation and metabolism, will need to be carried out in future studies along with microbial analysis. In urine, the identified metabolites, such as gallate, syringate, and vanillate can provide insight into microbial, metabolic, and feed parameters that cause changes depending on the feed rate. Additionally, it is thought that they can be used as potential biomarkers for further research on subacute ruminal acidosis.
The rumen environment is affected by many factors, such as diet type, feeding frequency, and the ratio of forage and concentrate [1]. In particular, the type of feed significantly affects rumen fermentation. The rumen microorganisms of ruminants decompose the feed and produce volatile fatty acid (VFA) as energy sources. In the forage fed, the buffering capacity of the diet is determined largely by the total chewing time, because the salivary buffer capacity of cows increases during chewing [2]. The effective fiber in forage is important in maintaining rumen function and stimulating the growth of fibrolytic bacteria, but the high content of indigestible fiber in corn stover has detrimental effects on nutrient utilization and digestibility, which depresses microbial protein synthesis and lowers nitrogen efficiency [3,4]. Compared to forage, concentrated feed consists of carbohydrates, such as starch, and can be rapidly decomposed in the rumen, resulting in a large amount of organic acid in the rumen, which can lead to acidosis.
The forage to concentrate ratio depends on the dietary ingredients and nutritional requirements of ruminant animals. Most previous studies have focused on the effects of different dietary forage to concentrate ratios on the bacterial community of Holstein heifers, energy utilization, growth rate, and feces microflora [5–7]. These processes are closely related to metabolic changes in the body. As cow health, growth, and metabolism are highly dependent on the production of metabolites in the rumen, a comprehensive analysis of the composition of rumen fluid can provide important insights into the role of rumen-diet interactions [8,9].
Previous studies of rumen on fed a high-concentrate diet have been observed in investigate feed efficiency, microbiota, and metabolites [10–12]. In a high concentrate diet (HCD), significantly increased the bacterial degradation product such as xanthine, uracil and amino acid such as leucine, glycine, and alanine in rumen metabolite. The relative abundance of cellulolytic bacteria and ciliates (Succinimonas, Fibrobacter, Polyplastron, and Ostracodinium) was linearly decreased according to increased concentrate diet level. Also, dairy cow on low pH in rumen had less backfat thickness and tended to have greater milk:feed ratio. Compared to the results of various studies on rumen, there are few studies on urine or feces that are relatively easy to sample in the field. Therefore, it is necessary to study of metabolite monitoring for change in various biofluids for dairy cows such as several previous studies [13,14].
We hypothesized that rumen acidosis caused by a HCD could be affect changes in urine and feces metabolite, and we thought that the difference could be used as research data for disease biomarkers. Therefore, the objective of this trial was to investigate the change of metabolites on the rumen fluid, urine, and feces of dairy cows brought about by different forage to concentrate ratios and exploration for potential biomarkers using proton nuclear magnetic resonance (NMR) analysis.
All experimental procedures including experimental animal maintenance and sample collection were conducted in accordance with guidelines from the committee of the National Institute of Animal Science, Rural Development Administration, Republic of Korea (Protocol Number: NIAS-2017-249). The study protocol was approved by the IACUC at National Institute of Animal Science.
Eight Holstein cows (average body weight, 598±90 kg; parity, 2±1; 167±18 days in milk) from the National Institute of Animal Science were used in this study. For the trial, cows were classified according to feed ratio factors. The cows used in the experiment were divided into two groups: normal concentrate diet (NCD, n = 4) and HCD (n = 4). All cows were fed an NCD group diet (10 kg; Italian ryegrass 80%: concentrate 20%) and HCD group diet (14.2 kg; Italian ryegrass 20%: concentrate 80%) twice a day at 1000 h and 1600 h on a dry matter (DM) basis. The chemical composition of the feed is presented in Supplementary Table S1.
The contents of DM (method No. 934.01), crude protein (CP, method No. 976.05), calcium (method No. 927.02), and phosphorus (method No. 3964.06) in Italian ryegrass and concentrate were assayed as described by Association of Official Analytical Communities methods [15]. The contents of neutral detergent fiber and acid detergent fiber in Italian ryegrass and concentrate were assayed as described by Van Soest et al [16]. The rumen pH of HCD group and NCD group was monitored for 6 consecutive hours after feeding in the morning every day. The NCD group confirmed that the appropriate pH range was maintained and HCD group make sure the pH was below 5.5 for more than 3 h. The experiment lasted 14 days, with first 7 days as the diet adaptation period [17].
Rumen fluid, urine, and feces samples were collected once for each animal, 4 h after the morning feeding on the last day of the experiment. Eight dairy cows were used for collection of rumen fluid through an oral stomach tube. The initially obtained rumen fluid (approximately 100 mL) was discarded to remove saliva and feed particle, and 200 mL of rumen fluid was collected in a conical tube. The pH of rumen fluid was analyzed immediately following collection using by pH meter (MP230, Mettler-Toledo, Columbus, OH, USA). Urine samples were collected after massage of the region underneath the vulva, in a 50 mL conical tube and immediately closed. Feces samples were collected directly through a clean anal area to the rectum of the animal in 50 mL conical tube. The collected samples were stored at −80°C until analysis.
The rumen fluid sample was thawed at 4°C and centrifuged at 12,902×g for 10 min to collect 300 μL of the supernatant. The reference material 2,2,3,3-d(4)-3-(Trimethylsilyl)propionic acid sodium salt (TSP) was dissolved in deuterium oxide (D2O) to make 300 μL of 0.4 mM and added with the prepared rumen fluid sample to make the 0.2 mM TSP concentration in the total mixed solution. The preparation of the rumen fluid sample was described elsewhere [14].
Urine samples added to 0.2 M sodium phosphate buffer (pH 7.0) were centrifuged at 14,000×g at 4°C for 10 min to collect 400 μL of supernatant. The supernatant was added to 230 μL of buffer and the pH was adjusted to 7.0±0.1. The 540 μL mixture was added to 5 mM TSP (60 μL) in D2O and the 0.5 mM TSP concentration of the 600 μL total solution. The preparation of urine samples was described elsewhere [14].
The feces sample thawed at 4°C was centrifuged at 14,000×g for 20 min to collect of the supernatant. The supernatant was recentrifuged at 12,902×g for 10 min and 400 μL sample was added to 140 μL sodium phosphate buffer (pH 7.0). The reference material TSP was dissolved in D2O to make 60 μL of 5 mM and added with the prepared fecal sample to make the 0.5 mM TSP concentration in the total mixed solution. The preparation of the fecal sample was described elsewhere [14].
The proton NMR spectra were acquired using AVANCE III HD Bruker spectrometer (Bruker BioSpin AG, Fallanden, Switzerland). To acquire spectra of the rumen, urine, and feces samples, the Bruker standard nuclear overhauser enhancement spectroscopy (NOESY) pre-saturation pulse sequence was used with the following parameters: temperature = 25°C, repetition number = 128, acquisition time = 2.0 s. The FID was acquired with a spectral width of 20 ppm and collected to 64k data points.
Metabolite qualitative and quantification were carried out by importing the analyzed spectral data into the Chenomx NMR suite 8.4 software (ChenomxInc, Edmonton, Canada). The spectra were manually baseline and phase using Chenomx processor 8.4 software. The spectral width was 10 ppm and was referenced to the TSP signal at 0 ppm. The metabolite databases used for classification were the bovine metabolome database (www.bovinedb.ca), livestock metabolome database (www.lmdb.ca), and the human metabolome database (www.hmdb.ca).
All metabolite data analyses were performed using by Metaboanalyst 4.0 (www.metaboanalyst.ca) [18]. The resulting data were normalization selected methods follow as raw wise normalization: normalization to constant sum; data transformation: Log10 normalization; data scaling: pareto scaling.
The between-group differential metabolites were analyzed using student’s t-test (p<0.05). The multivariate data analysis was performed principal component analysis (PCA), partial least squares-discriminant analysis (PLS-DA), orthogonal partial least squares-discriminant analysis (OPLS-DA) and variable importance in projection (VIP) was created based on PLS-DA results.
The classification of metabolites is shown in Figure 1. Original concentration of rumen fluid, urine, and feces are provided as supplementary information (Supplementary Tables S2–4). One hundred sixty metabolites were identified in the rumen fluid (four alcohols, three aliphatic acyclic compounds, seven amines, 15 amino acids, 13 benzoic acids, 27 carbohydrates, 18 carboxylic acids, three imidazolinones, three indoles, 15 lipids, 12 nucleosides and nucleotides, 18 organic acids, 21 others, and one pyridine).
In the urine samples, 177 metabolites (two alcohols, three aliphatic acyclic compounds, 10 amines, 18 amino acids, 18 benzoic acids, 24 carbohydrates, 23 carboxylic acids, three imidazolinones, three indoles, 21 lipids, four nucleosides and nucleotides, 15 organic acids, 32 others, and one pyridine) were identified.
In the feces samples, 188 metabolites (four alcohols, three aliphatic acylic compounds, seven amines, 22 amino acids, 16 benzoic acids, 30 carbohydrates, 28 carboxylic acids, four imidazolinones, three indoles, 19 lipids, six nucleosides and nucleotides, 19 organic acids, and 27 others) were identified.
The PCA score plot shows that the model interpretation rates for the HCD group and NCD group (Figure 2). The PCA revealed that PC 1 and 2 accounted for 41.5% and 16.7%, 28.6% and 18.7%, and 29.3% and 21.1% of the total variation in the rumen fluid, urine, and feces, respectively. The two groups of rumen fluid and urine samples were well separated, and samples from the same group were well aggregated together. On the other hand, the feces samples were not separated and showed similar tendencies between the two groups.
The PLS-DA is a versatile algorithm that can be used for predictive and descriptive modeling as well as for discriminative variable selection (Figure 3). In the score plot of PLS-DA, the NCD and HCD groups were discriminated with an R2 of 0.988, a Q2 of 0.870 in rumen fluid. In urine and feces was 0.940 and 0.551 and 0.791 and −0.6372, respectively. The R2 (coefficient of determination) explains how well the model fits the data, and R2 close to one is one of the necessary conditions for the model to be robust. The value of Q2 (cumulative prediction ability) indicates the ability of the model to predict new data, and a larger Q2 indicates that the model has good predictive performance.
An OPLS-DA supervised model was used to assess intergroup differences and permutation test (set permutation number 1,000) for statistical testing under the null hypothesis. In the rumen fluid and urine, R2Y, and Q2 were 0.999 and 0.947 and 0.999 and 0.792, respectively, whereas in the feces, R2Y = 0.489 and Q2 = 0.139 (Figure 4). Both the R2Y and Q2 values were greater than 0.4, indicating that the model was stable and reliable, except for the feces samples. A Q2 value of approximately 1 indicated that the OPLS-DA model had good predictability. The p-values of permutation for rumen fluid were 0.029 (R2Y) and 0.029 (Q2). In the p-value of urine and feces, R2Y and Q2 were 0.061 and 0.031 and 0.501 and 1. The VIP score >1.0 metabolite, and heat map of VIP scores are shown in Figure 5.
A univariate analysis using a t-test was conducted for rumen, urine, and feces metabolites for the NCD group and HCD group. The results of significantly (p<0.05) lower and higher metabolites between the two groups are shown in Tables 1 and 2, respectively. We identified a total of 64 differential metabolites in the rumen fluid, of which 15 had low concentrations and 49 had high concentrations in the HCD group compared to the NCD group. In the urine and feces samples, the metabolites with low and high concentrations were 14 and 16, and 2 and 2, respectively.
The rumen plays a central role in the efficiency of digestion in ruminants. The pattern of rumen metabolites shows the degradation products of rumen microorganisms, depending on the feed ratio. Non-fiber carbohydrates (NFCs), including starch, monosaccharides, oligosaccharides, and pectin, are rapidly decomposed by rumen microorganisms to produce VFAs [19]. A higher NFC ratio in feed increases the production of VFAs, resulting in a decrease in rumen pH [20]. Subacute rumen acidosis occurs when the rumen pH remains low between 5.8 and 5.5 at least 3 h/d [17]. Subacute ruminal acidosis leads to the destruction of bacteria composition balance due to the decrease in ruminal pH sensitive cellulolytic bacteria and the dominance of acid-resistant bacteria. The risk of ruminal acidosis causes various effects of economic loss such as low milk production and laminitis to endotoxin released from Streptococcus bovis [21].
In this study, the NFC content of NCD and HCD groups was 28.0% and 45.8% (DM %), respectively. According to a study by AlZahal et al [22], if the NFC content of the feed exceeds 40% of the DM standard, it is possible to reduce the rumen pH to less than 5.6 for 5.0 h/d.
The PCA and PLS-DA analyses showed a clear separation between rumen fluid and urine metabolites, except for feces metabolites, owing to the different diets offered in the present study, thereby representing a significant difference in metabolic changes.
In particular, metabolites related to carbohydrates and amino acids were significantly altered. We know that the feed ratio of the HCD group contained a lot of starch and polysaccharides were rapidly decomposed into glucose in the rumen. Among the carbohydrate metabolites, those that were significantly higher in the HCD group were glucose, arabinose, ribose, glucoronate, and xylose. In particular, xylose and arabinose account for up to 80% of the carbohydrates that comprise the hemicellulose of forage and cereals [23]. This is attributed to the fact that there are more soluble components in the concentrated feed. Glucuronate is a sugar acid derived from glucose, and its sixth carbon atom is oxidized to a carboxylic acid [24]. Glucuronate is a xylan degradation product and a source of pyruvate necessary for metabolic processes. In particular, glucuronate is an important growth substrate for b316, which plays a key role in the degradation of plant polysaccharides, producing butyrate as an end-product [25]. Leucine is an essential amino acid for mammals as a substrate for protein synthesis and is considered an efficient nutrient signal that regulates protein synthesis [26]. Histidine is an essential amino acid that cannot be metabolically synthesized and must therefore be absorbed from the diet. Histidine is used through bacterial metabolism to degrade glutamate, ammonia, and single carbon compounds [27]. 3-Methylhistidine is a metabolite of protein and energy metabolism, increased levels of which are observed in the rumen when the animal is fed a diet high in rumen-degradable protein [28]. In blood and urine, 3-Methylhistidine levels are used as biomarkers of skeletal muscle degradation and muscle protein turnover in animals [29]. Succinate was significantly higher in the NCD group, which had a high rate of forage. Many pure cultures of rumen bacteria form succinate as a final fermentation product in the rumen; however, this does not accumulate in the rumen [30]. Added succinate is rapidly converted to propionate in the rumen by washed rumen bacteria [31].
Urine is a liquid that excretes water-soluble by-products produced through metabolic pathways in animals. It has much to do with nitrogen circulation in the body. Urea is produced in the liver by the ureagenesis of ammonia, which is absorbed through the rumen wall. Colmenero et al [32] reported that the urinary level of urea is affected by the CP content of the feed. The increase in urea in the HCD group was similar to that reported in previous studies wherein the amount of urea-N excreted increased with the increase in the CP intake, and the correlation between the two was confirmed [33]. Amino acids, including alanine and glycine are produced by transamination in muscle and other tissues, and transport nitrogen for urea production through amino acid catabolism in the liver. The amino acid net flux of the liver represents the sum of unidirectional removal and release, and in the case of amino acids that can be released from the liver, the indication of unidirectional removal may be greater than the measured value of the net flux [34]. The relationship between the alanine and the high-concentrate diet groups observed in this study can be explained. Trimethylamine-N-oxide is synthesized from the oxidation of trimethylamine and is correlated with dietary choline. Soy, whole grains, and vegetables are sources of choline. Interestingly, our results showed that the concentration of forage-derived phenolic compounds (such as vanillate, syringate, gallate, and hippurate) was high in the NCD group. However, there was no significant difference between the groups. The anaerobic degradation of lignin-derived aromatic metabolites has been studied in various research fields [35]. Consequently, we confirmed the existence of gallate, vanillate, and syringate as degradation intermediates and lignin-derived methoxylated monoaromatics. The observed high values in three metabolites (vanillate, syringate, and gallate) could be interpreted as being a result of feeding a high forage ratio. Hippurate is a metabolite commonly identified in the urine of mammals and is related to the intake of phenolic compounds. Phenylalanine, chlorogenic acid, and catechin are metabolized to produce benzoic acid, which is then transferred to the mitochondria to produce hippurate [36]. Creatine plays an important role in the energy response through its conversion creatine phosphate via phosphorylation related to energy transfer, generating new ATP [35]. Creatine can accumulate in the body of animals through de novo synthesis and diet [37], and is mainly synthesized in the liver using glycine, arginine, and methionine, and transferred to tissue through the blood. Residual creatine is excreted through urine. It is thought to be significantly higher in HCD group than in NCD group due to the excretion of creatine, which is not used as an energy source in the body due to the high feeding of concentrated feed. Carnitine is required for the transport of long-chain fatty acids from the cytoplasm to the matrix compartment of the mitochondria for energy production, and is also involved in a variety of important physiological processes, including ketogenesis, thermogenesis, lipolysis, and potential nitrogen metabolism [38]. The urinary carnitine concentration was eight times higher in the HCD group than in the NCD group. Urinary carnitine is derived from the blood and can be used as a predictor of a high concentration of fatty acids that must be used in the body. The choline in feed is degraded by rumen microorganisms by approximately 80%, and metabolic choline is supplied by endogenous synthesis via the phosphatidylethanolamine N-methyltransferase pathway [39].
In this study, metabolites in the rumen fluid, urine, and feces of dairy cows with different feed ratios were screened as non-targets using proton NMR. The metabolites in the rumen fluid changed according to the feed rate. These included glucose, xylose, and maltose related to carbohydrate metabolism, and leucine, alanine, and isoleucine related to protein metabolism, which was found to be highly concentration in the HCD group. As HCD group is related to rumen microbial changes, research on different metabolites such as glucuronate, acetylsalicylate, histidine, and O-Acetylcarnitine, which are related to bacterial degradation and metabolism, will need to be carried out in future studies along with microbial analysis. In urine, muscle metabolism and microbial-derived metabolites such as creatine, alanine, and xanthine were identified. In addition, the identified metabolites, such as gallate, syringate, and vanillate originated from the NCD group. These metabolites can provide insight into microbial, metabolic, and feed parameters that cause changes depending on the feed rate; additionally, it is thought that they can be used as potential biomarkers for further research.
Notes
SUPPLEMENTARY MATERIAL
Supplementary file is available from: https://doi.org/10.5713/ab.22.0124
Supplementary Table S1. The formulation and chemical composition of experiment diet
ab-22-0124-suppl1.pdf
Supplementary Table S2. Concentrations of the rumen fluid metabolite by 1H-NMR analysis (μM, Median±interquartile range, n = 4)
ab-22-0124-suppl2.pdf
Supplementary Table S3. Concentrations of the urine metabolite by 1H-NMR analysis (μM, Median±interquartile range, n = 4)
ab-22-0124-suppl3.pdf
Supplementary Table S4. Concentrations of the feces metabolite by 1H-NMR analysis (μM, Median±interquartile range, n = 4)
ab-22-0124-suppl4.pdf
Figure 1
The classification of metabolite on rumen fluid, urine, and feces to the detectable metabolome. Identified metabolites are categorized according to chemical class (bovine metabolome database and livestock metabolome database) and the number of metabolites detected by proton nuclear magnetic resonance.
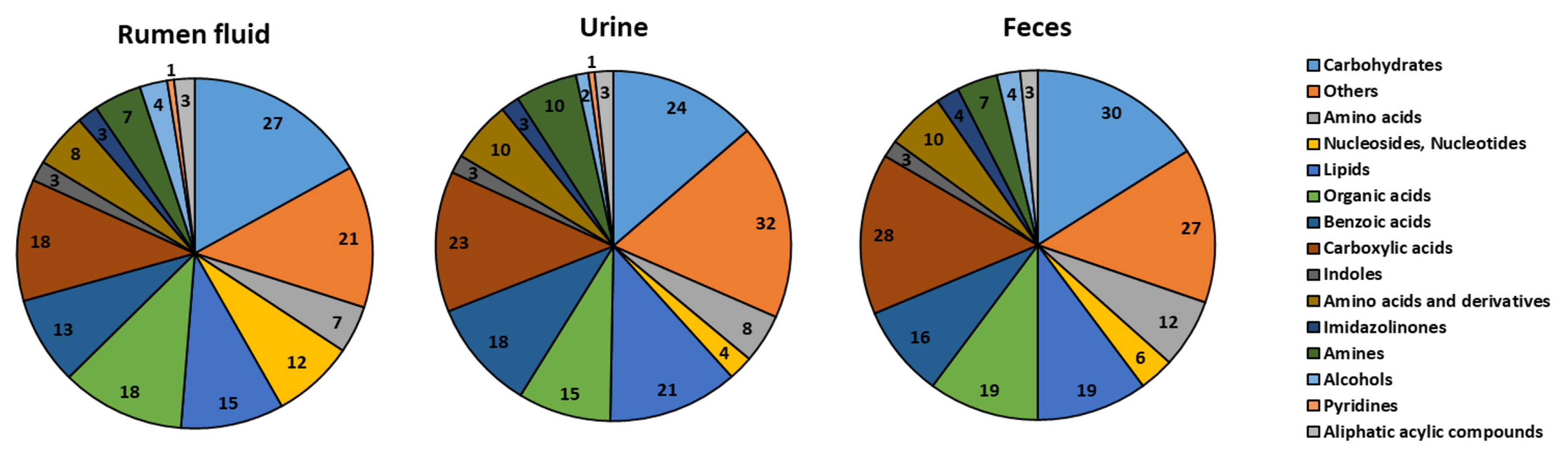
Figure 2
Principal component analysis (PCA) of the (A) rumen fluid, (B) urine, and (C) feces metabolite. The PCA plot distinguishes the metabolic profiles in cows fed diets based on high concentrate diet (HCD, red dots) vs normal concentrate diet (NCD, green dots). Ellipse represents 95% confidence interval.
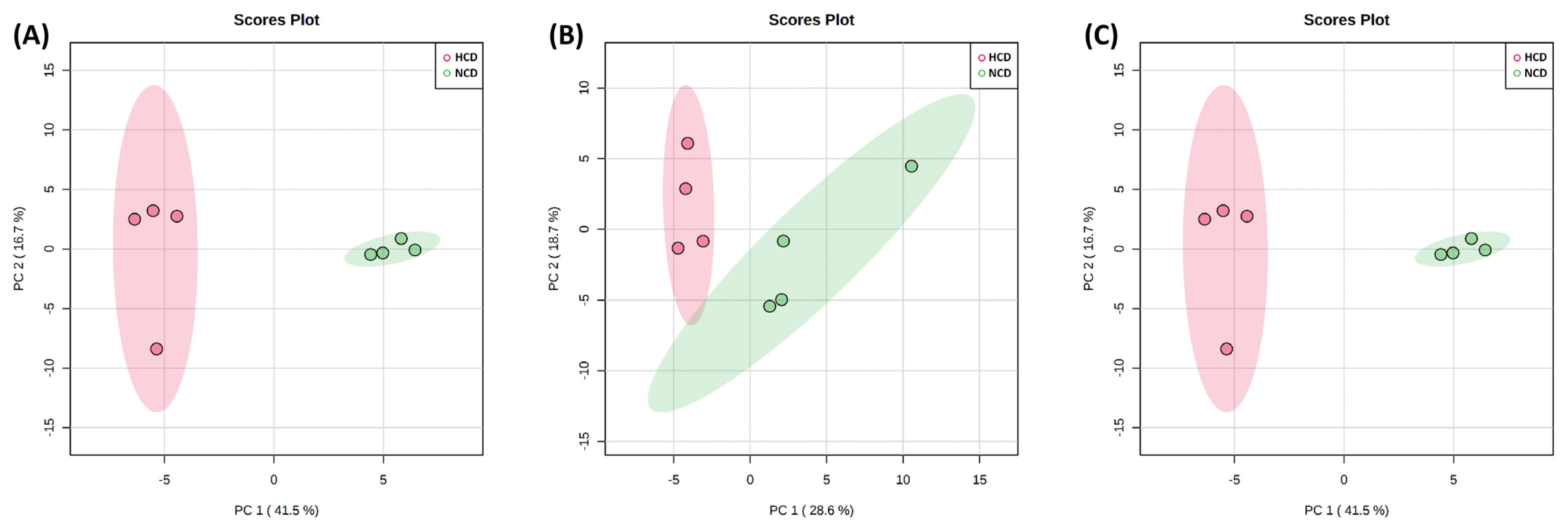
Figure 3
Partial least squares-discriminant analysis (PLS-DA) of the (A) rumen fluid, (B) urine, and (C) feces metabolite. The PLS-DA plot distinguishes the metabolic profiles in cows fed diets based on high concentrate diet (HCD, red dots) vs normal concentrate diet (NCD, green dots). Ellipse represents 95% confidence interval.

Figure 4
Orthogonal partial least squares-discriminant analysis (OPLS-DA) of the (A) rumen fluid, (B) urine, and (C) feces metabolite. The OPLS-DA plot distinguishes the metabolic profiles in cows fed diets based on high concentrate diet (HCD, red dots) vs normal concentrate diet (NCD, green dots). Ellipse represents 95% confidence interval.
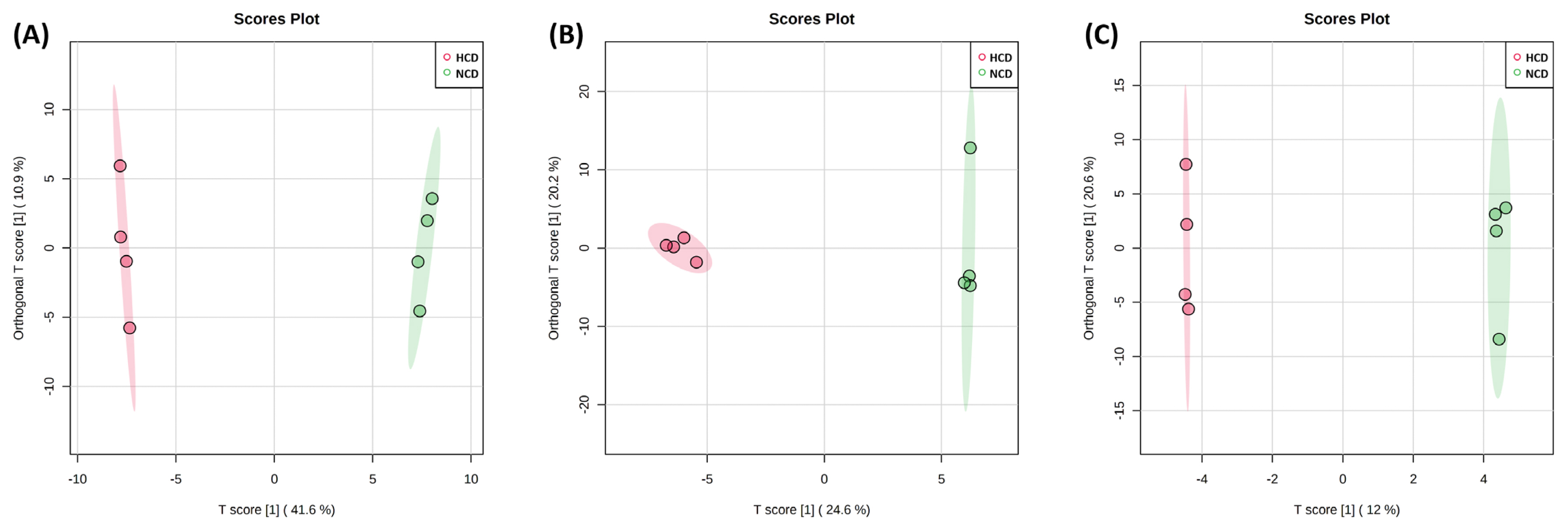
Figure 5
The top 30 metabolites of variable importance in projection (VIP) scores of metabolite in (A) rumen fluid, (B) urine, and (C) feces in cows fed diets based on high concentrate diet (HCD) vs normal concentrate diet (NCD). The selected metabolite were those with VIP>1.0 based on the parial least squares-discriminant analysis model. The red and blue colors of the heat map on the right indicate the abundance of each metabolite from high to low.
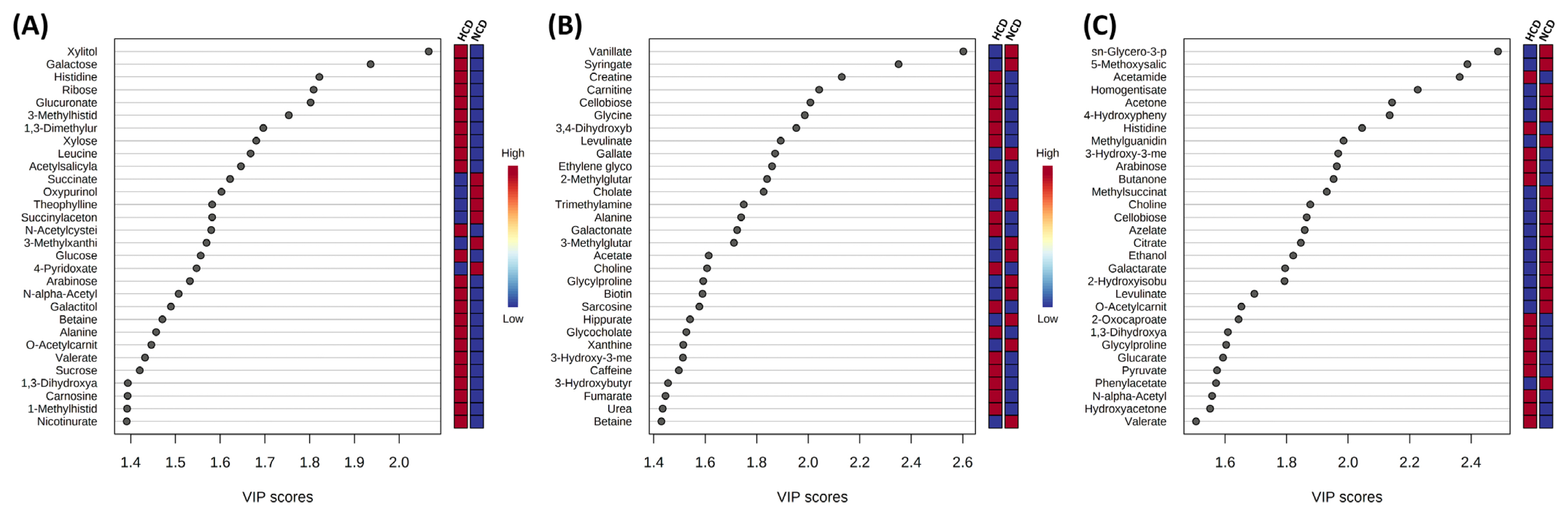
Table 1
The statistically low metabolites in high concentrate diet (HCD) compared to normal concentrate diet (NCD)
| Metabolite1) | FC2) | p-value | VIP3) | Metabolite | FC | p-value | VIP |
|---|---|---|---|---|---|---|---|
| Rumen fluid | |||||||
| Methylamine | 0.34 | 0.047 | 1.29 | Oxypurinol | 0.08 | <0.001 | 1.60 |
| Succinate | 0.08 | <0.001 | 1.62 | 4-Pyridoxate | 0.11 | <0.001 | 1.54 |
| UDP-N-Acetylglucosamine | 0.23 | 0.030 | 1.00 | 1,7-Dimethylxanthine | 0.14 | 0.005 | 1.29 |
| Succinylacetone | 0.08 | <0.001 | 1.58 | Ibuprofen | 0.22 | 0.042 | 0.98 |
| Ferulate | 0.14 | 0.014 | 1.23 | 3-Phenylpropionate | 0.38 | 0.003 | 0.98 |
| Gluconate | 0.18 | 0.050 | 1.01 | Acetate | 0.70 | <0.001 | 0.61 |
| Urine | |||||||
| Methanol | 0.31 | 0.039 | 1.42 | 3-Methylglutarate | 0.19 | 0.014 | 1.71 |
| Trimethylamine N-oxide | 0.17 | <0.001 | 1.74 | Thymol | 0.34 | 0.019 | 1.27 |
| Hippurate | 0.25 | <0.001 | 1.54 | Xanthine | 0.25 | <0.001 | 1.51 |
| Vanillate | 0.02 | <0.001 | 2.60 | Succinylacetone | 0.33 | 0.034 | 1.16 |
| Gallate | 0.12 | <0.001 | 1.87 | Betaine | 0.13 | 0.049 | 1.42 |
| Syringate | 0.16 | 0.021 | 2.35 | N-Methylhydantoin | 0.42 | 0.008 | 1.15 |
| Glycylproline | 0.21 | 0.001 | 1.59 | trans-Aconitate | 0.40 | 0.006 | 1.15 |
| Feces | |||||||
| 5-Methoxysalicylate | 0.19 | 0.045 | 2.38 | 4-Hydroxyphenyllactate | 0.20 | 0.016 | 2.13 |
Table 2
The statistically high metabolites in high concentrate diet (HCD) compared to normal concentrate diet (NCD)
| Metabolite1) | FC2) | p-value | VIP3) | Metabolite | FC | p-value | VIP |
|---|---|---|---|---|---|---|---|
| Rumen fluid | |||||||
| Isopropanol | 3.87 | 0.024 | 1.20 | Glycylproline | 3.31 | 0.042 | 1.01 |
| Kynurenine | 2.57 | 0.013 | 0.89 | 3-Hydroxyphenylacetate | 3.53 | 0.011 | 1.10 |
| Carnosine | 5.77 | 0.007 | 1.39 | 3-Hydroxyisovalerate | 3.73 | 0.009 | 1.15 |
| Anserine | 4.59 | 0.002 | 1.27 | N-Carbamoylaspartate | 4.00 | 0.007 | 1.23 |
| 1-Methylhistidine | 7.32 | 0.003 | 1.39 | cis-Aconitate | 5.22 | 0.004 | 1.35 |
| 3-Methylhistidine | 15.94 | 0.001 | 1.75 | Homovanillate | 6.08 | 0.007 | 1.39 |
| Histidine | 18.85 | <0.001 | 1.82 | N-Acetylcysteine | 8.51 | 0.004 | 1.58 |
| Isoleucine | 5.05 | 0.001 | 1.26 | N-alpha-Acetyllysine | 8.57 | 0.004 | 1.50 |
| Alanine | 6.42 | 0.004 | 1.45 | 5-Hydroxyindole-3-acetate | 3.27 | 0.003 | 1.11 |
| Leucine | 11.63 | 0.001 | 1.66 | 3-Hydroxy-3-methylglutarate | 3.12 | 0.020 | 1.03 |
| 4-Hydroxyphenylacetate | 2.84 | 0.001 | 1.04 | O-Acetylcarnitine | 9.96 | 0.033 | 1.44 |
| 3,4-Dihydroxybenzeneacetate | 3.91 | <0.001 | 1.21 | NAD | 4.71 | <0.001 | 1.25 |
| o-Cresol | 4.74 | 0.023 | 1.32 | Butyrate | 1.57 | <0.001 | 0.69 |
| 3-Hydroxymandelate | 5.01 | <0.001 | 1.32 | N-Nitrosodimethylamine | 2.93 | 0.015 | 1.03 |
| Acetylsalicylate | 10.72 | 0.002 | 1.64 | Salicylate | 3.83 | <0.001 | 1.20 |
| Maltose | 5.64 | 0.002 | 1.30 | Nicotinate | 4.63 | <0.001 | 1.29 |
| 1,3-Dihydroxyacetone | 5.93 | <0.001 | 1.39 | Phenylacetate | 5.17 | 0.003 | 1.24 |
| Glucose | 8.93 | <0.001 | 1.55 | Nicotinurate | 6.19 | <0.001 | 1.39 |
| Galactitol | 9.71 | 0.001 | 1.48 | Valerate | 6.63 | 0.004 | 1.43 |
| Arabinose | 9.84 | 0.003 | 1.53 | 2-Hydroxyphenylacetate | 3.87 | 0.029 | 0.98 |
| Ribose | 17.27 | 0.001 | 1.80 | Desaminotyrosine | 4.16 | <0.001 | 1.24 |
| Glucuronate | 18.33 | <0.001 | 1.80 | Caffeine | 4.18 | 0.018 | 1.21 |
| Xylose | 22.69 | 0.006 | 1.68 | Betaine | 5.96 | 0.013 | 1.47 |
| Galactose | 34.71 | <0.001 | 1.93 | 1,3-Dimethylurate | 17.57 | 0.003 | 1.69 |
| Xylitol | 46.96 | <0.001 | 2.06 | ||||
| Urine | |||||||
| Urea | 3.24 | <0.001 | 1.43 | Hydroxyacetone | 2.52 | 0.047 | 1.35 |
| Sarcosine | 4.19 | 0.002 | 1.57 | Choline | 4.88 | 0.027 | 1.60 |
| N-Phenylacetylglycine | 3.14 | 0.037 | 1.39 | Cholate | 5.30 | 0.030 | 1.82 |
| Creatine | 48.02 | 0.029 | 2.13 | 2-Methylglutarate | 6.00 | 0.014 | 1.84 |
| Alanine | 5.88 | <0.001 | 1.73 | Carnitine | 8.58 | 0.006 | 2.04 |
| Glycine | 27.40 | 0.023 | 1.98 | Ethylene glycol | 9.56 | 0.006 | 1.85 |
| 3,4-Dihydroxybenzeneacetate | 9.79 | 0.002 | 1.95 | Levulinate | 10.42 | 0.003 | 1.89 |
| Galactonate | 6.70 | 0.025 | 1.72 | Cellobiose | 7.49 | 0.016 | 2.00 |
| Feces | |||||||
| Histidine | 4.51 | 0.020 | 2.04 | 2-Oxocaproate | 2.82 | 0.050 | 1.64 |
REFERENCES
1. Li R, Teng Z, Lang C, et al. Effect of different forage-to-concentrate ratios on ruminal bacterial structure and real-time methane production in sheep. PLoS One 2019; 14:e0214777
https://doi.org/10.1371/journal.pone.0214777



2. Bailey CB, Balch CC. Saliva secretion and its relation to feeding in cattle: 1. The composition and rate of secretion of parotid saliva in a small steer. Br J Nutr 1961; 15:371–82.
https://doi.org/10.1079/BJN19610047


3. Cook DE, Combs DK, Doane PH, Cecava MJ, Hall MB. The effects on digestibility and ruminal measures of chemically treated corn stover as a partial replacement for grain in dairy diets. J Dairy Sci 2016; 99:6342–51.
https://doi.org/10.3168/jds.2015-10403


4. Lascano GJ, Koch LE, Heinrichs AJ. Precision-feeding dairy heifers a high rumen-degradable protein diet with different proportions of dietary fiber and forage-to-concentrate ratios. J Dairy Sci 2016; 99:7175–90.
https://doi.org/10.3168/jds.2016-11190


5. Bi Y, Zeng S, Zhang R, Diao Q, Tu Y. Effects of dietary energy levels on rumen bacterial community composition in Holstein heifers under the same forage to concentrate ratio condition. BMC Microbiol 2018; 18:69
https://doi.org/10.1186/s12866-018-1213-9



6. Bremer J. Carnitine--metabolism and functions. Physiol Rev 1983; 63:1420–80.
https://doi.org/10.1152/physrev.1983.63.4.1420


7. Claffey NA, Fahey AG, Gkarane V, Moloney AP, Monahan FJ, Diskin MG. Effect of forage to concentrate ratio and duration of feeding on growth and feed conversion efficiency of male lambs. Transl Anim Sci 2018; 2:419–27.
https://doi.org/10.1093/tas/txy071



8. Xia C, Muhammad A, Niu W, et al. Effects of dietary forage to concentrate ratio and wildrye length on nutrient intake, digestibility, plasma metabolites, ruminal fermentation and fecal microflora of male Chinese Holstein calves. J Integr Agric 2018; 17:415–27.
https://doi.org/10.1016/S2095-3119(17)61779-9

9. Saleem F, Bouatra S, Guo AC, et al. The bovine ruminal fluid metabolome. Metabolomics 2013; 9:360–78.
https://doi.org/10.1007/s11306-012-0458-9

10. Ametaj BN, Zebeli Q, Saleem F, et al. Metabolomics reveals unhealthy alterations in rumen metabolism with increased proportion of cereal grain in the diet of dairy cows. Metabolomics 2010; 6:583–94.
https://doi.org/10.1007/s11306-010-0227-6

11. Zhang R, Zhu W, Jiang L, Mao S. Comparative metabolome analysis of ruminal changes in Holstein dairy cows fed low- or high-concentrate diets. Metabolomics 2017; 13:84
https://doi.org/10.1007/s11306-017-1204-0

12. Zhang J, Shi H, Wang Y, et al. Effect of dietary forage to concentrate ratios on dynamic profile changes and interactions of ruminal microbiota and metabolites in Holstein heifers. Front Microbiol 2017; 8:2206
https://doi.org/10.3389/fmicb.2017.02206



13. Eom JS, Kim ET, Kim HS, et al. Metabolomics comparison of rumen fluid and milk in dairy cattle using proton nuclear magnetic resonance spectroscopy. Anim Biosci 2021; 34:213–22.
https://doi.org/10.5713/ajas.20.0197



14. Kim HS, Kim ET, Eom JS, et al. Exploration of metabolite profiles in the biofluids of dairy cows by proton nuclear magnetic resonance analysis. PLoS One 2021; 16:e0246290
https://doi.org/10.1371/journal.pone.0246290



15. AOAC International. Official methods of analysis. Seventeen editionGaithersburg, MD, USA: Association of Official Analytical Chemists; 2003.
16. Van Soest PJ, van Robertson JB, Lewis B. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J Dairy Sci 1991; 74:3583–97.
https://doi.org/10.3168/jds.S0022-0302(91)78551-2


17. Gozho GN, Plaizier JC, Krause DO, Kennedy AD, Wittenberg KM. Subacute ruminal acidosis induces ruminal lipopolysaccharide endotoxin release and triggers an inflammatory response. J Dairy Sci 2005; 88:1399–403.
https://doi.org/10.3168/jds.S0022-0302(05)72807-1


18. Chong J, Wishart DS, Xia J. Using metaboanalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr Protoc Bioinform 2019; 68:e86
https://doi.org/10.1002/cpbi.86

19. Polakova K, Kudrna V, Kodes A, Hucko B, Mudrik Z. Non-structural carbohydrates in the nutrition of high-yielding dairy cows during a transition period. Czech J Anim Sci 2010; 55:468–78.
https://doi.org/10.17221/873-CJAS

20. AlZahal O, Kebreab E, France J, Froetschel M, McBride BW. Ruminal temperature may aid in the detection of subacute ruminal acidosis. J Dairy Sci 2008; 91:202–7.
https://doi.org/10.3168/jds.2007-0535


21. Mackie RI, Gilchrist FMC. Changes in lactate-producing and lactate-utilizing bacteria in relation to pH in the rumen of sheep during stepwise adaptation to a high-concentrate diet. Appl Environ Microbiol 1979; 38:422–30.
https://doi.org/10.1128/aem.38.3.422-430.1979



22. AlZahal O, Kebreab E, France J, McBride BW. A mathematical approach to predicting biological values from ruminal pH measurements. J Dairy Sci 2007; 90:3777–85.
https://doi.org/10.3168/jds.2006-534


23. Wilkie KCB. The hemicelluloses of grasses and cereals. Advances in Carbohydrate Chemistry and Biochemistry. Elsevier; 1979. p. 215–64.
https://doi.org/10.1016/S0065-2318(08)60237-1

24. Dutton GJ. The mechanism of glucuronide formation. Biochem Pharmacol 1961; 6:65–71.
https://doi.org/10.1016/0006-2952(61)90074-0


25. Kelly WJ, Leahy SC, Altermann E, et al. The glycobiome of the rumen bacterium Butyrivibrio proteoclasticus B316T highlights adaptation to a polysaccharide-rich environment. PLoS One 2010; 5:e11942
https://doi.org/10.1371/journal.pone.0011942



26. Davis T, Suryawan A, Orellana RA, Fiorotto ML. Leucine acts as a nutrient signal to stimulate protein synthesis. J Anim Sci 2010; 88:E-Suppl22
27. Bergen WG, Purser DB, Cline JH. Determination of limiting amino acids of rumen-isolated microbial proteins fed to rat. J Dairy Sci 1968; 51:1698–700.
https://doi.org/10.3168/jds.S0022-0302(68)87255-8


28. Kaufman JD, Pohler KG, Mulliniks JT, Ríus AG. Lowering rumen-degradable and rumen-undegradable protein improved amino acid metabolism and energy utilization in lactating dairy cows exposed to heat stress. J Dairy Sci 2018; 101:386–95.
https://doi.org/10.3168/jds.2017-13341


29. Kamiya M, Kamiya Y, Tanaka M, Shioya S. Milk protein production and plasma 3-methylhistidine concentration in lactating Holstein cows exposed to high ambient temperatures. Asian-Australas J Anim Sci 2006; 19:1159–63.
https://doi.org/10.5713/ajas.2006.1159

30. Russell JB, Hespell RB. Microbial rumen fermentation. J Dairy Sci 1981; 64:1153–69.
https://doi.org/10.3168/jds.S0022-0302(81)82694-X


31. Blackburn TH, Hungate RE. Succinic acid turnover and propionate production in the bovine rumen. Appl Microbiol 1963; 11:132–5.
https://doi.org/10.1128/am.11.2.132-135.1963



32. Colmenero JJO, Broderick GA. Effect of dietary crude protein concentration on milk production and nitrogen utilization in lactating dairy cows. J Dairy Sci 2006; 89:1704–12.
https://doi.org/10.3168/jds.S0022-0302(06)72238-X


33. Jardstedt M, Hessle A, Nørgaard P, Richardt W, Nadeau E. Feed intake and urinary excretion of nitrogen and purine derivatives in pregnant suckler cows fed alternative roughage-based diets. Livest Sci 2017; 202:82–8.
https://doi.org/10.1016/j.livsci.2017.05.026

34. Lindsay DB. Amino acids as energy sources. Proc Nutr Soc 1980; 39:53–9.
https://doi.org/10.1079/PNS19800008


35. Kato S, Chino K, Kamimura N, Masai E, Yumoto I, Kamagata Y. Methanogenic degradation of lignin-derived monoaromatic compounds by microbial enrichments from rice paddy field soil. Sci Rep 2015; 5:14295
https://doi.org/10.1038/srep14295



36. Lees HJ, Swann JR, Wilson ID, Nicholson JK, Holmes E. Hippurate: The natural history of a mammalian-microbial cometabolite. J Proteome Res 2013; 12:1527–46.
https://doi.org/10.1021/pr300900b


37. Wyss M, Kaddurah-Daouk R. Creatine and creatinine metabolism. Physiol Rev 2000; 80:1107–213.
https://doi.org/10.1152/physrev.2000.80.3.1107


38. Borum PR. Role of carnitine during development. Can J Physiol Pharmacol 1985; 63:571–6.
https://doi.org/10.1139/y85-097


39. Goselink RMA, Van Baal J, Widjaja HCA, et al. Effect of rumen-protected choline supplementation on liver and adipose gene expression during the transition period in dairy cattle. J Dairy Sci 2013; 96:1102–16.
https://doi.org/10.3168/jds.2012-5396








 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Supplement1
Supplement1 Print
Print





