 |
 |
| Anim Biosci > Volume 35(7); 2022 > Article |
|
Abstract
Objective
In this study, we aimed to identify long non-coding RNAs (lncRNAs) that play important roles in starvation stress, analyze their functions, and discover potential molecular targets to alleviate starvation stress to provide a theoretical reference for subsequent in-depth research.
Methods
We generated a piglet starvation stress animal model. Nine Yorkshire weaned piglets were randomly divided into a long-term starvation stress group (starved for 72 h), short-term starvation stress group (starved for 48 h), and the control group. LncRNA libraries were constructed using high-throughput sequencing of piglet ileums.
Results
We obtained 11,792 lncRNAs, among which, 2,500 lncRNAs were novel. In total, 509 differentially expressed (DE)lncRNAs were identified in this study. Target genes of DElncRNAs were predicted via cis and trans interactions, and functional and pathway analyses were performed. Gene ontology functions and Kyoto encyclopedia of genes and genomes analysis revealed that lncRNA-targeted genes mainly participated in metabolic pathways, cellular processes, immune system processes, digestive systems, and transport activities. To reveal the mechanism underlying starvation stress, the interaction network between lncRNAs and their targets was constructed based on 26 DElncRNAs and 72 DEmRNAs. We performed an interaction network analysis of 121 DElncRNA–DEmRNA pairs with a Pearson correlation coefficient greater than 0.99.
Conclusion
We found that MSTRG.19894.13, MSTRG.16726.3, and MSTRG.12176.1 might play important roles in starvation stress. This study not only generated a library of enriched lncRNAs in piglets, but its outcomes also provide a strong foundation to screen key lncRNAs involved in starvation stress and a reference for subsequent in-depth research.
The World Health Organization (WHO) reports that since 1975, the number of obese people in the world has grown nearly three-fold, and currently, nearly 20 million adults are overweight or obese [1]. Obesity not only affects one’s physical appearance but also causes various diseases [2] that burden the economy and the medical and health infrastructure. At present, popular methods of weight loss include dieting, targeted drug therapy, and surgical liposuction. Weight loss based on long-term fasting has significant effects, but it is not healthy [3]. Short-term starvation stress might be beneficial for cardiometabolic health by decreasing insulin resistance, blood pressure, and oxidative stress [4]. Long-term starvation stress can lead to a deficit of effective nutrients, increase levels of corticosterone, the stress hormone, and inhibit cardiovascular function [5].
The intestine is the main site of the digestion, absorption, and metabolism of nutrients in the body. The host intestinal mucosa directly or indirectly participates in metabolism, and its function is closely related to metabolic diseases such as obesity, diabetes, and allergies; moreover, it plays an active role in immune regulation [6]. Studies have reported that starvation can cause changes in the intestinal structure, damage the intestinal barrier, decrease the villus height-to-crypt depth (V/C) ratio, and dysregulate the intestinal flora, which eventually leads to metabolic disorders [7].
Long non-coding RNAs (lncRNAs) are a class of non-coding RNAs longer than 200 nucleotides that are widely present in the nucleus and cytoplasm [8]. LncRNAs participate in the regulation of various biological processes (BPs), such as gene expression, transcriptional activation, cell cycle regulation, and disease occurrence [9]. Recent studies have surmised that an increasing number of lncRNAs play important roles in maintaining intestinal epithelial homeostasis, promoting the proliferation and apoptosis of intestinal epithelial cells, and regulating the intestinal barrier [10–12]. For example, lncRNA H19 promotes the expression of miR-675-5p and miR-675-3p, which downregulate the levels of tight junction proteins 1 (TJP1, also called ZO-1) and E-cadherin, destroying the intestinal epithelial barrier [10]. In mice with starvation-induced mucosal atrophy, lncRNA uc.173 promotes the proliferation of intestinal epithelial cells by reducing the expression of miR-195 [11]. Transcriptome sequencing performed on ulcerative colitis tissue revealed lncRNA BC012900 overexpression. Furthermore, the overexpression of this lncRNA promotes intestinal epithelial cell apoptosis [12].
Owing to the availability of genetic information, easy genetic modification, and low cost of rearing, mice are the preferred in vivo models for studying human diseases [5,13]. However, significant differences exist in metabolic and physiological characteristics between humans and mice, which prevents researchers from applying findings from murine studies to humans for metabolism-related disease prevention and intervention strategies [14]. In contrast, pigs are very similar to humans in terms of metabolic characteristics and organ development; therefore, pigs are an ideal animal model for human energy metabolism research [15]. Thus, in this study, we subjected weaned piglets to different periods of starvation and used their ileum tissue for RNA sequencing analysis. In this study, we aimed to identify lncRNAs that have important functions in starvation stress, analyze their functions, and discover potential molecular targets for alleviating starvation stress, to provide a theoretical reference for subsequent in-depth research.
This research strictly adhered to the principles of animal use prescribed by the China Laboratory Animal Science Association. The study was approved by the Animal Ethics Committee of Shanxi Agricultural University (Taigu, China). The approval number for the Ethics Committee agreement was SXAU-EAW-2018P002005. The animals were humanely sacrificed as necessary to ameliorate suffering.
Yorkshire piglets were bred at the animal experiment station of Shanxi Agricultural University in accordance with the National Research Council (NRC) [16]. The test animals were from three litters of half-sibling piglets of the same birth age. On the weaning day, three boars with similar body weights (25 d-old, 5.96±0.15 kg) were selected from each litter, and nine weaned Yorkshire piglets were divided into three groups according to the block design principle. The piglets were randomly divided into a long-term starvation stress group (72 h, abbreviated as IHT-72), short-term starvation stress group (48 h, abbreviated as IHT-48), and a normal group (control group, abbreviated as ICT). The piglets were sacrificed, and ileum samples, which connected to the ileocecal ligament, were collected. The samples were washed with phosphate-buffered saline and immediately frozen in liquid nitrogen until RNA extraction.
The ileum samples (ICT, IHT-48, and IHT-72) were analyzed following hematoxylin and eosin (H&E) staining (Bosterbio, Wuhan, Hubei, China). The tissue samples were fixed in 4% paraformaldehyde for 24 h and processed using routine histological methods. Subsequently, 7 μm-thick sections were cut using a Leica RM2265 (Leica, Wetzlar, Germany) and stained. Three images were acquired (Olympus, Tokyo, Japan) for each section, and three sections were selected for each piglet. The lengths of intestinal villi were calculated and counted using SPSS ver.22.0 (IBM Corp., Armonk, NY, USA).
Total RNA was extracted from the nine libraries using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. RNA integrity was detected by performing 1% agarose gel electrophoresis, and the purity and concentration were determined using an ND-2000 nucleic acid-protein analyzer (Nanodrop Technologies, Winooski, VT, USA). The ratio of absorbances at 260 nm and 280 nm was determined, with values ranging from 1.8 to 2.0, indicating an RNA concentration of approximately 1,000 ng/μL. After total RNA was extracted, the rRNAs were removed to retain mRNAs and ncRNAs. The cDNA library was generated according to the instructions of the TruSeqRapid Duo cBot Sample Loading kit (Illumina, San Diego, CA, USA) and sequenced using an Illumina HiSeqTM 4000 by Gene Denovo Biotechnology Co. (Guangzhou, China).
To obtain high-quality clean reads, reads were filtered using fastp [17] (version 0.18.0). Data were cleaned by removing reads containing adapters, those containing more than 10% poly(N), and low-quality reads (containing more than 50% low-quality [Q-value≤20] bases) from the raw data. The short-read sequence alignment tool Bowtie2 [18] (version 2.2.8) was used to map the read sequence to the ribosomal RNA (rRNA) database. An index of the reference genome (Sscrofa11.1 [GCF_000003025.6]) was built, and paired-end clean reads were mapped to the reference genome using HISAT2 (version 2.1.0). The rRNA database uses the Nucleotide Sequence Database. Transcripts were reconstructed using Stringtie software (version 1.3.4), which together with HISAT2 allowed us to identify new genes and new splice variants of known genes.
The lncRNAs were screened according to a previously described process (SI Appendix, Figure S1). From the remaining transcripts that overlapped (>1 bp) with pig protein-coding genes, transcripts <200 bp and single-exon transcripts were removed. The coding potential calculator (CPC) (version 0.9-r2) and Coding-Non-Coding-Index (CNCI) (version 2) tools were used to assess the coding potential of the remaining transcripts, and transcripts with a CPC score >0 and CNCI score >0 were removed. The intersection of both non-protein-coding potential results was considered lncRNAs.
Differential expression of RNA and lncRNAs among different groups was analyzed using DESeq2 software. In addition, different samples in the same group were compared with edgeR. Transcripts with a false discovery rate (FDR) less than 0.05 and an absolute fold change ≥2 were considered differentially expressed. Differentially expressed coding RNAs were subjected to enrichment analysis based on gene ontology (GO) functions and Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis.
Sequence alignment, location, and sequence feature analyses were conducted using the NCBI (https://www.ncbi.nlm.nih.gov), Ensembl (http://asia.ensembl.org/index.html), and UCSC (https://genome.ucsc.edu) websites. GO and KEGG pathway enrichment analyses were conducted using DAVID (http://david.niaid.nih.gov). We performed gene set enrichment analysis (GSEA) using GSEA [19] and MSigDB [19] software to identify whether a set of genes in specific pathways was associated with significant differences between two groups.
lncRNAs regulate mRNA expression levels via cis and trans interactions. Target genes within 10 kb of differentially expressed lncRNAs (DElncRNA) were selected to explore cis-regulation, and DElncRNA–mRNA target gene pairs with a Pearson correlation coefficient greater than 0.99 were selected to explore co-expression regulation. DElncRNA–mRNAs were visualized using Cytoscape v3.8.2.
The extracted total RNA was diluted to 500 ng/μL and reverse-transcribed according to the instructions of the PrimeScript RT reagent kit with gDNA Eraser (TaKaRa Bio, Dalian, China). real time quantitative polymerase chain reaction (RT-qPCR) was performed on a CFX96 Real-Time PCR detection system (Bio-Rad, Hercules, CA, USA) using a SYBR Green RT-qPCR kit (TaKaRa Bio, China). 18S rRNA was used as an internal reference gene, and the sequences of primers used are listed in Supplementary Table S1. Three technical replicates were performed for each sample, along with a non-template negative control and a negative control without reverse transcriptase. The RT-qPCR cycling parameters were as follows: pre-denaturation at 95°C for 30 s, followed by 40 cycles of denaturation at 95°C for 30 s and annealing/extension at 60°C for 30 s. Melting curve analysis comprised a denaturation step at 95°C for 30 s, 60°C for 1 min, and 95°C for 30 s.
RT-qPCR results were used to calculate the relative expression levels of the target genes using the 2−ΔΔCt method. Statistical analyses were performed using SPSS ver.22.0 (IBM Corp., USA). Differences in gene expression among the different groups were identified using analysis of variance using Duncan’s test for significance. Pearson correlation analysis was used to determine the relationship between lncRNA expression and mRNA expression and to verify RNA-seq and RT-qPCR results. Graphpad version 8.0 (GraphPad Software, San Diego, CA, USA) was used to draw histograms. All tests were two-tailed, and a p-value <0.05 was considered a significant difference.
The morphological comparison of the piglet ilea under different treatments (Figure 1A) revealed that an increase in starvation time corresponded with shortening of the ileal villi (Figure 1B) and increases in the crypt depth (Figure 1C) and ratio of villus height to crypt depth (Figure 1D). In summary, as the starvation stress time increased, the ileum tissue damage increased.
The raw reads of each sample were between 11.1 and 14.7 billion. After excluding the aptamer sequence and low-quality reads, 773,785,766 clean reads were obtained from the nine RNA-Seq datasets (Table 1). The Q30 of each sample was ≥91.96%, the N percentage was 0.00%, and the guanine-cytosine (GC) percentage of each library was between 44.20% and 46.22%. These data demonstrated that the sequencing had high reliability. After removing the rRNA reads from the alignment, they were compared to the pig reference genome, and 96.67% to 95.03% reads were successfully mapped. Stringties was used to reconstruct transcripts, which were compared with the known transcripts recorded in the Ensembl database. Subsequently, 11,792 lncRNAs and 63,682 mRNAs were obtained, including 2,500 novel lncRNAs and 69 novel mRNAs (SI Appendix, Table S1). We determined the coding potential of the novel lncRNAs using the prediction software CNCI and CPC2 (Figure 2A).
According to the relative position of lncRNAs and protein-coding genes in the genome, lncRNAs can be divided into five categories, intergenic lncRNAs, bidirectional lncRNAs, intronic lncRNAs, antisense lncRNAs, and sense overlapping lncRNAs. Among them, intergenic lncRNAs were the most common in our study, with 7,333 accounting for approximately 62% of lncRNAs. Intronic lncRNAs were the least abundant, with a total of 272, accounting for approximately 2% of lncRNAs (Figure 2B).
The average number of exons in lncRNAs was 3.90±2.65, which was significantly less than the average number of exons in mRNAs (13.66±11.64) (Figure 2C). The average exon length in lncRNAs was 1.03±2.46 kb, which was significantly longer than the exon length in mRNAs (0.31±0.80 kb) (Figure 2D). The average transcript length of lncRNAs was 4.01± 5.62 kb, which was shorter than the transcript length of mRNAs (4.23±3.15 kb) (Figure 2E). The average open reading frame (ORF) length of lncRNAs was 0.29±0.30 kb, which was significantly shorter than the ORF length of mRNAs (2.15±1.99 kb) (Figure 2F). These results indicate that piglet lncRNAs have fewer but longer exons, longer transcripts, and longer ORFs than mRNA transcripts.
To assess the reliability and operational stability of the test results, we performed a Pearson analysis of lncRNA expression in each sample. The correlation coefficients between the samples were between 0.9804 and 0.9977 (SI Appendix, Figure S2). The samples in each treatment group showed a very high correlation with each other. Among them, the correlation between the replicates within the ICT and IHT-72 groups was greater than 0.99. However, the second sample in the IHT-48 group had a low correlation with the other two samples, although both values were greater than 0.98. A comparison of the correlation between groups found that ICT and IHT-48 groups had a better correlation, and both of these treatment groups had a weaker correlation with the IHT-72 group (Figure 3A). In the principal component analysis (PCA), the ICT and IHT-48 groups were located relatively closely but were situated relatively far from the IHT-72 group (Figure 3B). This indicates that differences in the ileum can be observed after 72 h of starvation stress.
The fragments per kilobase of transcript per million mapped reads (FKPM) values of lncRNAs (average 2.76) were significantly lower than those of the coding genes (average 14.82), including 23 highly expressed lncRNAs (over 100) and 259 moderately expressed lncRNAs (between 10 and 100). Based on an FDR of 5% and a q-value <0.05, 509 DElncRNAs (Figure 4A, 4B) and 3,319 DEmRNAs (Figure 4C, 4D) were identified by pairwise comparisons among the three treatment groups. There were nine DElncRNAs between ICT and IHT-48, among which, the expression level of one was upregulated, whereas those of eight were downregulated (Figure 4E). There were 275 DElncRNAs between IHT-48 and IHT-72, of which, the expression levels of 24 were upregulated and those of 151 were downregulated (Figure 4F). There were 396 DElncRNAs between ICT and IHT-72, of which, expression levels of 116 were upregulated and those of 280 were downregulated (Figure 4G). The trends in lncRNAs and mRNAs were similar. A Venn diagram showed that DElncRNAs and DEmRNAs were mainly concentrated in IHT-48 vs IHT-72 and ICT vs IHT-72 comparisons (Figure 4B, 4D), indicating that the piglet ileum underwent significant changes after 72 h of starvation. These results were verified by GSEA of DEmRNAs. Compared to the control animals and animals subjected to 48 h of starvation, animals subjected to 72 h of starvation showed decreased metabolism. This decrease affected pathways such as glycolysis, gluconeogenesis, the tricarboxylic acid cycle (TCA cycle), and fructose and mannose metabolic pathways. It also impacted pyruvate metabolism, as well as vitamin digestion and absorption. Furthermore, this treatment increased inflammation-related cells and diseases, such as cell adhesion molecules (CAMs), Th1/Th2 cell differentiation, inflammatory bowel disease, and malaria (SI Appendix, Figure S3).
Numerous studies have indicated that lncRNAs exert either positive or negative effects to regulate the expression of neighboring protein-coding genes [20,21]. Therefore, we extracted the information related to the location of differential lncRNAs from the transcription file and porcine genome database (http://genome-asia.ucsc.edu ) and selected mRNAs located within 10 kb upstream or downstream of these lncRNAs. We screened 59 DElncRNAs and 57 DEmRNAs and obtained 64 pairs of DElncRNA–DEmRNAs (SI Appendix, Table S2). GO analysis revealed that the DElncRNAs–DEmRNAs were enriched in BPs, cell components (CCs), and molecular functions (MFs). Among these, 21 BPs, 15 CCs, and eight MFs were involved (Figure 5A), and the proportion of genes enriched in terms such as metabolic processes, developmental processes, cell parts, catalytic activity, and nucleic acid binding was higher. To clarify the specific signaling pathways affected by the targets, KEGG analysis was conducted. KEGG analysis results showed that the target genes were enriched in 97 pathways (Figure 5B). Among the top 20 significantly enriched pathways, those related to energy metabolism were most enriched. These included seven items including metabolic pathways, fructose and mannose metabolism, and fatty acid metabolism, among others. The pathways related to drug absorption and metabolism, including drug metabolism - cytochrome P450, drug metabolism by other enzymes, and chemical carcinogenesis were the second most abundant. Four disease-related pathways, namely involved in hepatocellular carcinoma, prostate cancer, fluid shear stress in atherosclerosis, and cancer were also enriched. The cis-regulation analysis showed that after the piglets were subjected to starvation stress, significant changes occurred in energy homeostasis, metabolism, drug absorption, and disease occurrence.
In trans-regulation, the function of lncRNA is not linked to the mRNA location but to the co-expressed mRNA. We screened 447 DElncRNAs and 2,601 DEmRNAs and obtained 38,265 pairs of DElncRNAs–DEmRNAs. GO analysis revealed that 26 BPs were involved, and 19 CC sand 11 MFs were enriched (Figure 6A). Similar to the cis-regulated GO results, these DElncRNAs were determined to have important functions in regulating metabolic processes, cellular components, nucleic acid binding, and transcription factor activity. KEGG analysis showed that the target genes were enriched in 328 pathways (Figure 6B). Most of the top 20 pathways with significant enrichment were related to energy metabolism, including metabolic pathways, protein digestion and absorption, mineral absorption, fat digestion and absorption, and vitamin digestion and absorption, among others. However, enrichment was also observed in other pathways, such as drug metabolism-cytochrome P450, arrhythmogenic right ventricular cardiomyopathy, and tight junctions. These enrichment results were similar to the enrichment results of cis-regulation, indicating that the piglets exhibited significant changes to their metabolism and susceptibility to disease during starvation stress.
Cytoscape was used to map DElncRNAs/DEmRNAs with a Pearson correlation coefficient absolute value greater than 0.99 (p<0.01; SI Appendix, Figure S4). The results showed a total of 1,000 interactions among 212 DElncRNAs and 599 DEmRNAs. There were 998 positive interactions (red lines) and two negative (black lines) interactions among pairs within the network, and most DElncRNA–DEmRNA pairs were positively correlated. To more clearly identify lncRNAs that play a key role in hunger stress, 121 pairs of DElncRNAs–DEmRNAs with both GO and KEGG enrichment results and ranking in the top 15 in node number were selected (including 26 DElncRNAs and 72 DEmRNAs), as shown in Figure 6C. All of these networks had positive interactions (red lines). Among them, lncRNA XR_002336098.1 had the most co-expressed mRNAs (12 mRNAs), whereas complement component 4 binding protein alpha (C4BPA) mRNA had the most co-expressed lncRNAs (7 lncRNAs). The functions of DElncRNAs and DEmRNAs in the interaction network were mainly enriched in metabolism and disease. For example, MSTRG.19894.13 participates in metabolism by regulating the expression of serine dehydratase like (SDSL), UDP-glucose 6-dehydrogenase (UGDH), and xylulokinase (XYLB), among other genes, and MSTRG.16726.3 participates in human diseases by regulating the expression of complement C3 (C3), complement factor H (CFH), and glutathione S-transferase omega 1 (GSTO1), among other genes. In the interaction network, MSTRG.12176.1 had more interactive pairs and was expressed at higher levels, which suggested that it might be the key lncRNA in the ileum of piglets subjected to long-term starvation stress. Further, it regulates metabolism by modulating the expression of genes such as UDP-galactose-4-epimerase (GALE) and pyridoxal kinase (PDXK) and can participate in human diseases by regulating the expression of genes such as caudal type homeobox 2 (CDX2) and phospholipase C beta 3 (PLCB3).
To verify the reliability of the RNA sequencing data, three DElncRNAs (XR_001302663.2, MSTRG.16726.3, and MSTRG.19894.13; Figure. 7A) and three DEmRNAs (C3, C4BPA, and GSTO1; Figure 7B) were randomly selected. Based on NCBI (https://www.ncbi.nlm.nih.gov) and UCSC (https://genome.ucsc.edu) reference sequences, primers were designed using Primer 5.0 to validate these lncRNAs and mRNAs (SI Appendix, Table S3). The RT-qPCR results showed that the expression profiles of XR_001302663.2, C3, C4BPA, and GSTO1 were consistent with the sequencing results, and the expression profiles of MSTRG.16726.3 and MSTRG.19894.13 were similar to the sequencing results (Figure 7A). The expression levels of these lncRNAs and mRNAs were comparable in ICT and IHT-48 groups and were significantly higher than those in the IHT-72 group.
Despite the continuous increase in food production in modern times, starvation continues in various parts of the world either due to poverty or geological disasters. In contrast, several individuals starve voluntarily; for example, more people are inclined to adopt improper ways to lose weight by fasting for long durations [3]. The pig was used in this experiment as an ideal model to study human metabolic diseases [15]. In this study, we analyzed the transcriptional changes in piglets subjected to starvation stress via high-throughput sequencing and revealed the damage caused by starvation stress to the body from the perspective of lncRNA expression.
Genome-wide studies have revealed that lncRNAs play an important role in pigs [22]. However, few studies have been conducted on the expression of starvation stress-related lncRNA expression in pigs. In this study, we established a piglet model of starvation stress and found that the intestine is one of the key tissues damaged by starvation stress. Illumina HiSeqTM 4000 high-throughput sequencing was used to construct differential expression profiles of starvation stress-related lncRNAs in the piglet ileum. By testing the relationships between the samples, we found that at the transcriptome level, negligible differences were observed between the control group and the group starved for 48 h. However, compared with the ICT and IHT-48 groups, notable differences were observed in the group starved for 72 h. In our study, 72 h starvation inhibited glycolysis, gluconeogenesis, the TCA cycle, fructose and mannose metabolism, pyruvate metabolism, and vitamin digestion and absorption. This result suggests that long-term starvation leads to an insufficient energy supply, exhaustion of glycogen storage, and nutrient deficits. Furthermore, long-term starvation affects the body’s immunity and increases the risk of disease. During 72 h of starvation stress, mRNAs related to CAMs were overexpressed. The CAMs play an important role in the occurrence of diseases and are closely related to inflammation [23]. Further, 72 h starvation could induce Th1/Th2 cell differentiation and lead to inflammatory bowel disease and malaria. However, it might reduce the onset of diabetes, which could be related to the insufficient energy supply. In summary, prolonged starvation can cause irreversible damage to the body and at the same time provides a theoretical basis for the “golden 72 hours” associated with earthquake rescue.
A co-expression sub-network containing 26 DElncRNAs and 72 DEmRNAs was reconstructed to reveal the mechanisms underlying starvation stress and its effect. LncRNA MSTRG.19894.13 was determined to be an important lncRNA in the constructed network. C4BPA, dehydrogenase/reductase 4 (DHRS4), SDSL, UGDH, and XYLB were identified as the target genes of this lncRNA. C4BPA controls the classic complement activation pathway and interacts with RelA, a member of the nuclear factor kappa-B (NF-κB) family [24]. The expression of C4BPA is regulated by stress [24]. DHRS4 is a peroxisomal member of the short-chain dehydrogenase/reductase superfamily. Unlike that in its human counterpart, a mutation at Thr177 with the corresponding residue Asn stabilizes DHRS4 in pigs and prevents its cold-induced inactivation [25]. SDSL, also known as SDH2, exhibits low serine dehydratase and threonine dehydratase activities [26]. UGDH is a cytosolic enzyme that catalyzes the oxidation of UDP-glucose to UDP-glucuronic acid, thereby participating in the biosynthesis of glycosaminoglycans, such as hyaluronic acid, chondroitin sulfate, and heparan sulfate [27]. In patients with lung adenocarcinoma, the presence of UGDH in the nucleus is correlated with poorly differentiated cells and larger tumors and could indicate overall reduced survival [28]. XYLB encodes a xylulose kinase-like protein, and its forced overexpression is an effective strategy to improve xylose utilization and P(3HB) production in Burkholderia sacchari [29]. Our results showed that MSTRG. 19894.13 regulates the expression of C4BPA, DHRS4, SDSL, UGDH, and XYLB, and therefore, it might play an important role in regulating functions such as complement activation, enzyme activity, biosynthesis, and energy utilization.
The co-expressed target genes of MSTRG.16726.3 were determined to be aldo-keto reductase family 1 member A1 (AKR1A1), C3, CFH, GST01, MYB proto-oncogene (MYB), and polymeric immunoglobulin receptor (PIGR). Studies have shown that AKR1A1 can synthesize ascorbic acid in rodents [30] and is involved in the metabolism of gamma-hydroxybutyric acid in human liver cancer-derived HepG2 cells [30]. In contrast, C3 plays a central role in activation of the complement system and can be used as a biomarker for insulin resistance and cardiometabolic diseases [31]. CFH plays an important role in regulating complement activation and limits the effect of innate defense mechanisms against microbial infection [32]. GSTO1 is involved in the metabolism of xenobiotics and carcinogens, such as with cutaneous malignant melanoma [33]. Mutations in and the overexpression of MYB were first discovered in leukemia cells and were recently discovered in solid cancers [34]. MYB plays an important role in hematopoietic functions and tumorigenesis [34]. The circ-XPO4 of the small extracellular vesicles present in milk promotes the expression of PIGR in intestinal cells by inhibiting miR-221-5p, thereby increasing the level of intestinal SIgA [35]. In conclusion, MSTRG.16726.3 might play an important role in complement activation, the metabolism of exogenous and carcinogenic substances, resistance against pathogenic microorganisms, and improvements in intestinal immunity.
The following nine genes are targeted by MSTRG.12176.1: C-C motif chemokine ligand 25 (CCL25), CDX2, GALE, hepatocyte nuclear factor 4 gamma (HNF4G), PDXK, PLCB3, RAB27B, solute carrier family 2 member 5 (SLC2A5), and solute carrier family 34 member 3 (SLC34A3). CCL25, a member of the chemokine CC subfamily, has also been recognized as a thymus-expressed chemokine [36]. CCL25 and its receptor C-C motif chemokine receptor 9 (CCR9) are overexpressed in cancers such as leukemia, ovarian cancer, breast cancer, prostate cancer, liver cancer, lung cancer, and melanoma [36]. CDX2 is a major regulator of intestine-specific genes involved in cell growth and differentiation [37]. Decreased expression of CDX2 is associated with mucinous tumors, lymph node involvement, and high-grade tumors [37]. GALE encodes UDP-galactose-4-epimerase, and mutations in GALE result in epimerase-deficient galactosemia, also referred to as galactosemia type 3, a disease characterized by early onset cataracts, liver damage, deafness, and mental retardation [38]. HNF4G is overexpressed in colorectal cancer (CRC) and promotes CRC cell proliferation via the PI3K/AKT pathway by targeting G protein subunit gamma 12 (GNG12) and protein tyrosine kinase 2 (PTK2) [39]. PDXK encodes a pyridoxal kinase, which converts inactive B vitamins to the active cofactor pyridoxal 5′-phosphate (PLP) [40]. Hereditary polyneuropathy with optic atrophy can occur due to a PDXK variant leading to impaired vitamin B6 metabolism [40]. PLCB3 is involved in innate immunity, and studies have shown that silencing PLCB3 enhances the inflammatory signaling cascade of toll-like receptors [41]. RAB27B is an RAS oncogenic family member gene that regulates extracellular vesicle production in cells infected with Kaposi’s sarcoma-associated herpesvirus to promote cell survival and persistent infection [42]. The fructose transporter encoded by SLC2A5 is required for intestinal fructose absorption [43]. SLC2A5 expression is induced in the intestine and skeletal muscle of patients with type 2 diabetes and in certain cancers dependent on fructose metabolism [43]. In renal brush border cells, SLC34A3 is involved in transporting intracellular phosphate via sodium co-transport and contributes to the maintenance of inorganic phosphate concentrations in the kidneys [44]. Mutations in SLC34A3 can cause hereditary hypophosphatemic rickets with hypercalciuria [44]. Therefore, by acting on multiple target genes, MSTRG.12176.1 can play an important role in the starvation stress in piglets and affect energy metabolism, nutrient absorption, and disease occurrence and development.
In this study, 11,792 lncRNAs and 63,682 mRNAs were identified through transcriptome sequencing of ileum tissue obtained from pigs subjected to starvation stress. These included 2,500 novel lncRNAs, 509 DElncRNAs, and 3,319 DEmRNAs. The target genes of DElncRNAs were determined to be mainly involved in metabolic pathways, cellular processes, immune system processes, digestive systems, and transport activities, among others. A co-expression network induced by starvation stress in pigs was constructed, and the key lncRNAs were analyzed. However, the specific mechanism by which these lncRNAs regulate metabolism is unknown. Further studies are required to elucidate the molecular mechanisms linking lncRNAs to the regulation of starvation stress in pigs.
Notes
CONFLICT OF INTEREST
We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript. Gao PF is an employee of Inner Mongolia Mengniu Dairy (GROUP) CO., LTD.
FUNDING
This work was funded by the key research projects in agriculture of the Shanxi Province (201803D221022-1), the Program for Sanjin Scholar (Grant Nos. 2016 and 2017), the Fund for Shanxi 1331 Project (Grant No. 2017), the National Natural Science Foundation of China (Grant No. 31872336), and the Shanxi Agriculture University Science & Technology Innovation Fund (2017YJ07).
ACKNOWLEDGMENTS
We would like to thank all the staff of Gene Denovo Biotechnology Co. for technical help and Editage 360 ( www.editage.cn ) for English language editing during the preparation of this manuscript.
SUPPLEMENTARY MATERIAL
Supplementary file is available from: https://doi.org/10.5713/ab.21.0483
SI Appendix, Figure S1. Screening process for lncRNAs.
SI Appendix, Figure S2. Pearson correlation analysis between samples.
SI Appendix, Figure S3. GSEA enrichment analysis.
SI Appendix, Table S1. The transcript information statistics of ileum tissue of starvation-stressed pigs
SI Appendix, Table S2. Potential cis-regulation between porcine DELncRNA and its neighboring mRNAs
SI Appendix, Table S3. Primers used in this study
ab-21-0483-suppl.pdf
Figure 1
(A) Histological analysis of Sus scrofa ileum under starvation stress. ICT is the control, IHT-48 indicates piglets subjected to starvation stress for 48 h, and IHT-72 is piglets subjected to starvation stress for 72 h. (B-D) Histogram comparing the length of the villi, depth of the crypt, and villi/crypt ratio in the ileum among the two treatments and the control (A–C indicates p<0.01).
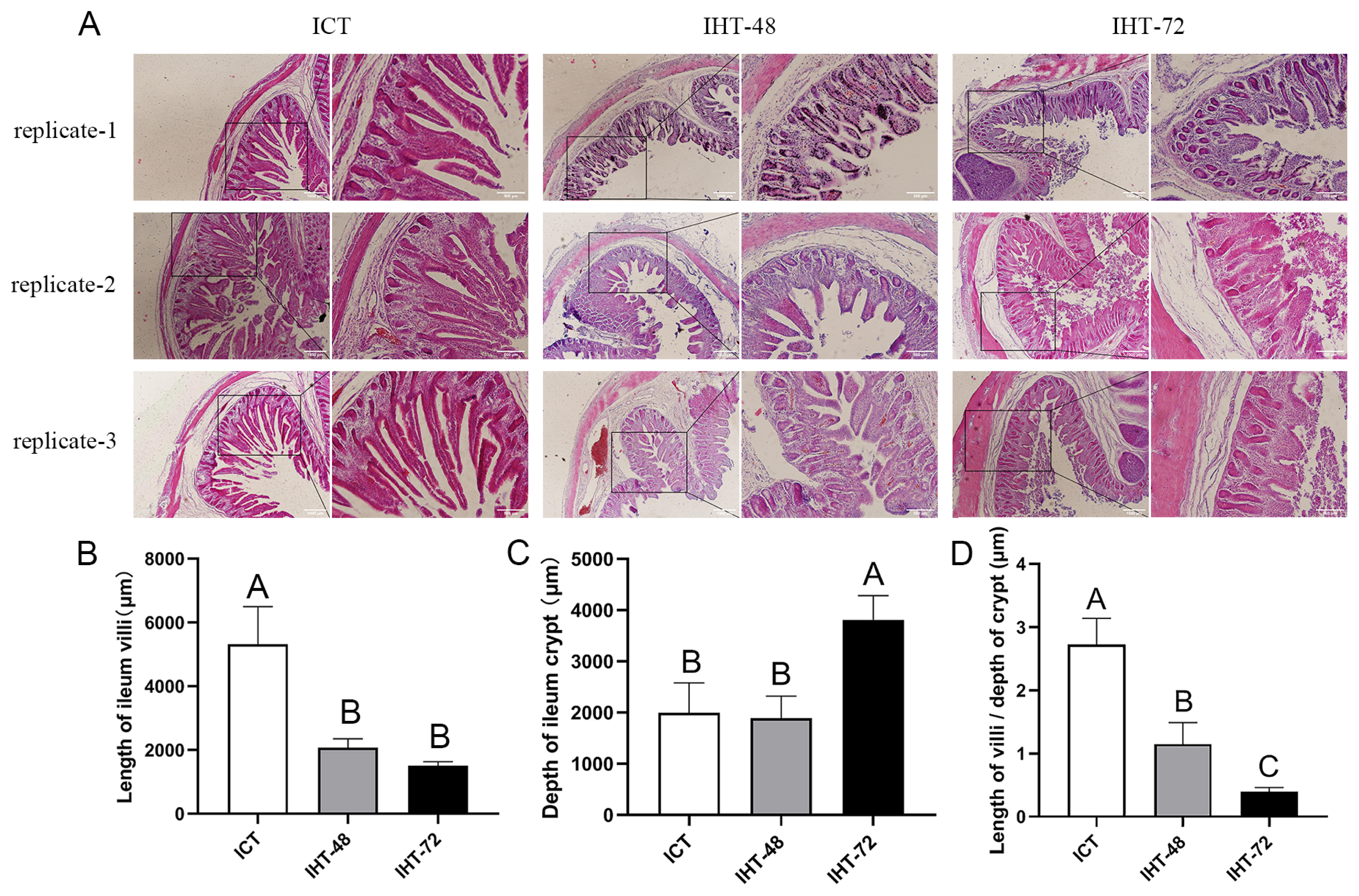
Figure 2
Identification and expression profile analysis of lncRNAs obtained from Sus scrofa ileum under starvation stress for 48 h and 72 h and control conditions. (A) Novel lncRNA-coding ability prediction. (B) Type of lncRNAs. (C) Number of lncRNA and mRNA exons. (D) Lengths of lncRNA and mRNA exons. (E) Length of lncRNA and mRNA transcripts. (F) Length of lncRNA and mRNA transcript ORFs. ORF, open reading frames.
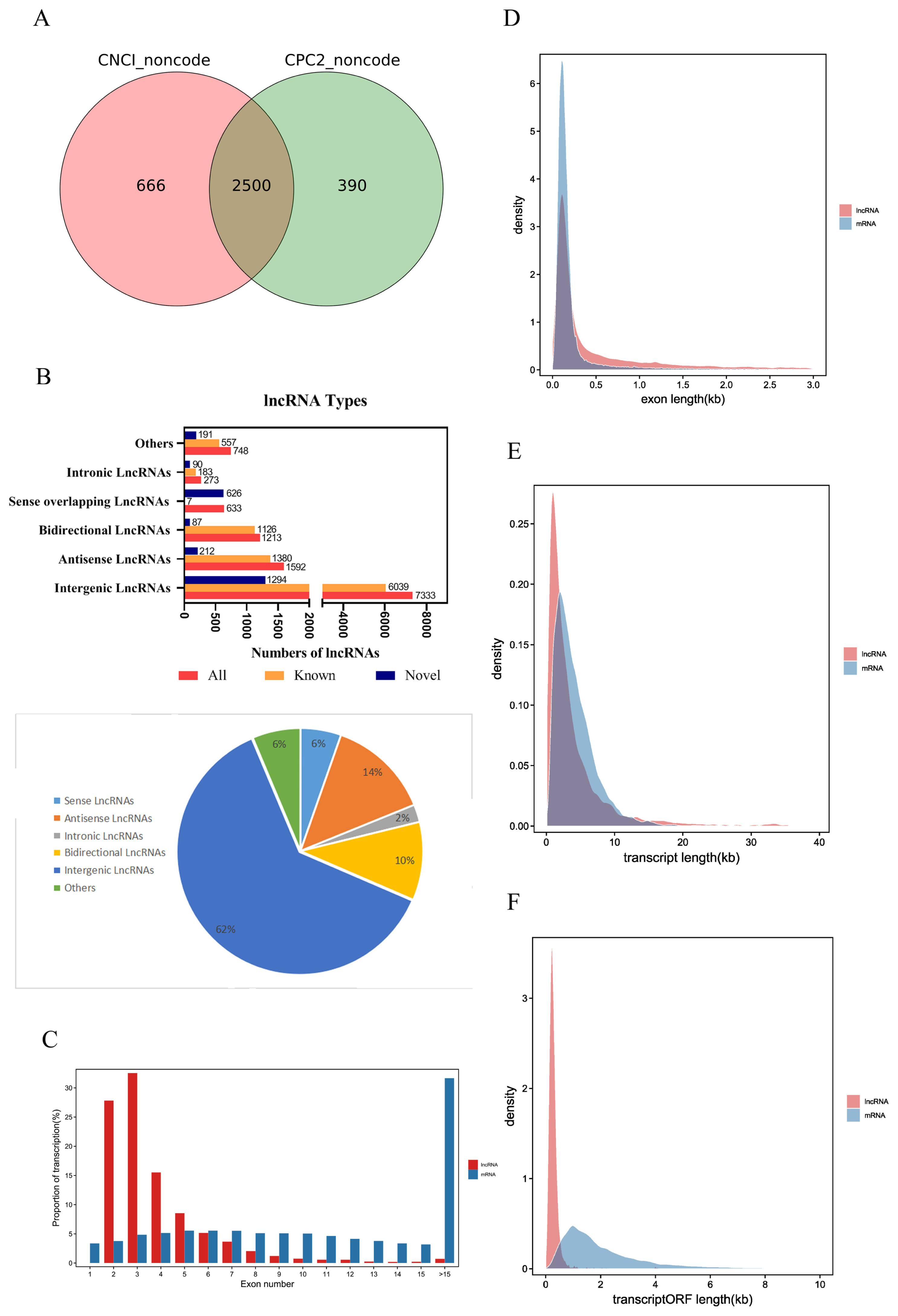
Figure 3
Sample correlation analysis of Sus scrofa ileum under starvation stress. ICT is the control, IHT-48 indicates piglets subjected to starvation stress for 48 h, and IHT-72 is piglets subjected to starvation stress for 72 h. (A) Correlation heat map. (B) PCA analysis. PCA, principal component analysis.
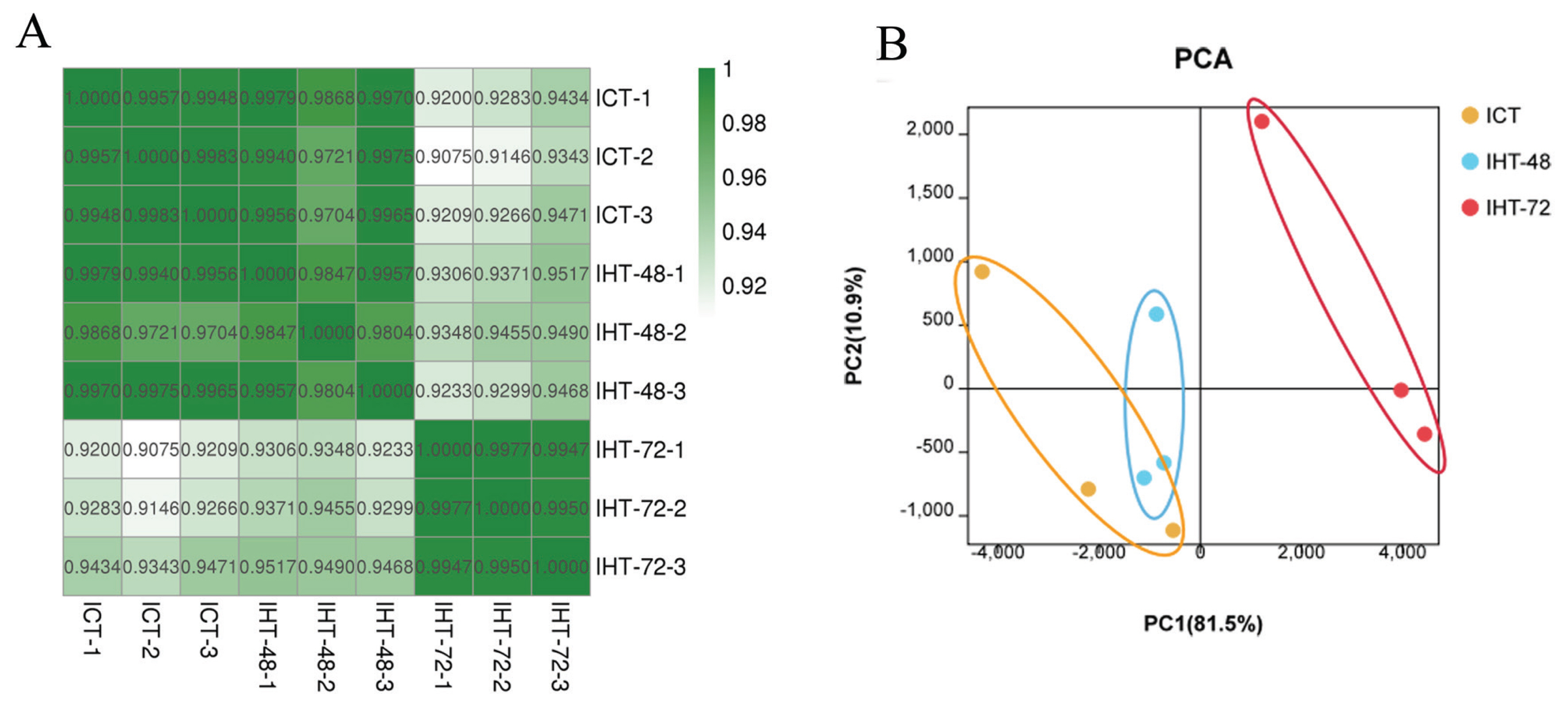
Figure 4
Analysis of differentially expressed (DE)lncRNAs and DEmRNAs of Sus scrofa ileum under starvation stress. ICT is the control, IHT-48 is piglets subjected to starvation stress for 48 h, and IHT-72 is piglets subjected to starvation stress for 72 h. (A) The bar graph shows the number of DElncRNAs. (B) Venn diagram showing DElncRNAs. (C) The bar graph shows the number of DEmRNAs. (D) Venn diagram of DEmRNAs. (E) Heat map of differentially expressed genes in ICT group and IHT-48 group (z-scores). (F) Heat map of differentially expressed genes in IHT-48 group and IHT-72 group (z-scores). (G) Heat map of differentially expressed genes in ICT group and IHT-72 group (z-scores).
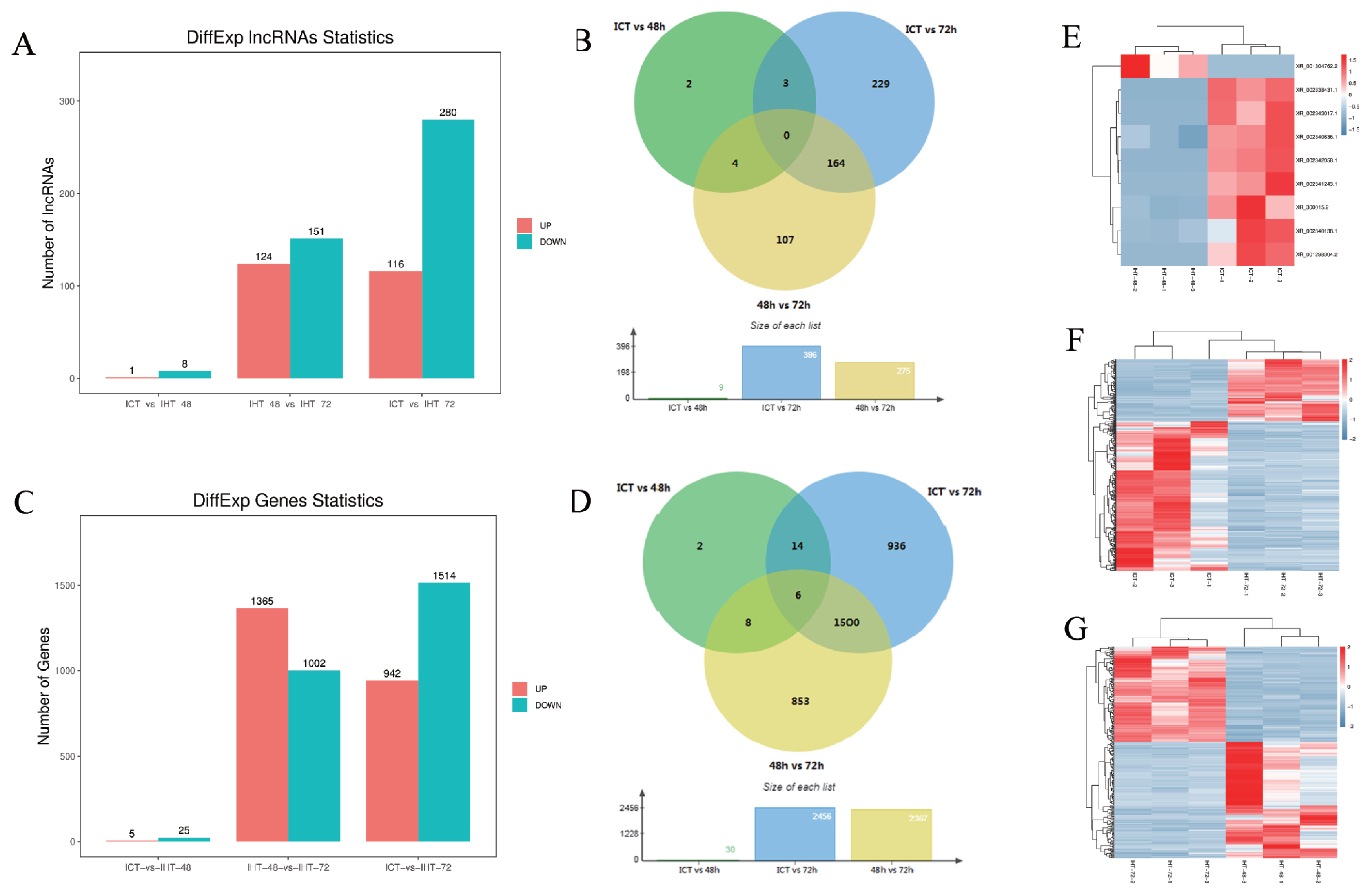
Figure 5
Potential cis-regulatory interaction between differentially expressed (DE)lncRNA and neighboring mRNAs in Sus scrofa ileum under starvation stress. (A) GO analysis. (B) KEGG analysis. GO, gene ontology; KEGG, Kyoto encyclopedia of genes and genomes.
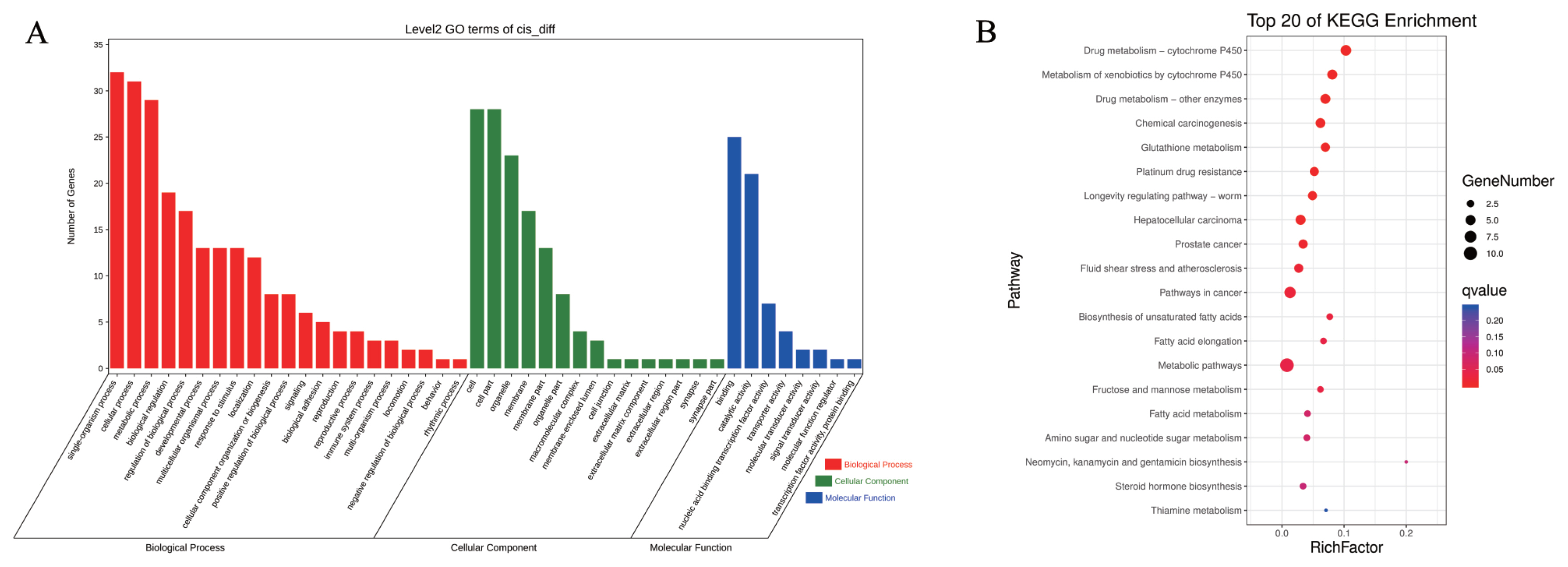
Figure 6
Potential trans-regulatory interactions between differentially expressed (DE)lncRNA and co-expressed mRNAs in Sus scrofa ileum under starvation stress. (A) GO analysis. (B) KEGG analysis. (C) Co-expression network analysis. GO, gene ontology; KEGG, Kyoto encyclopedia of genes and genomes.
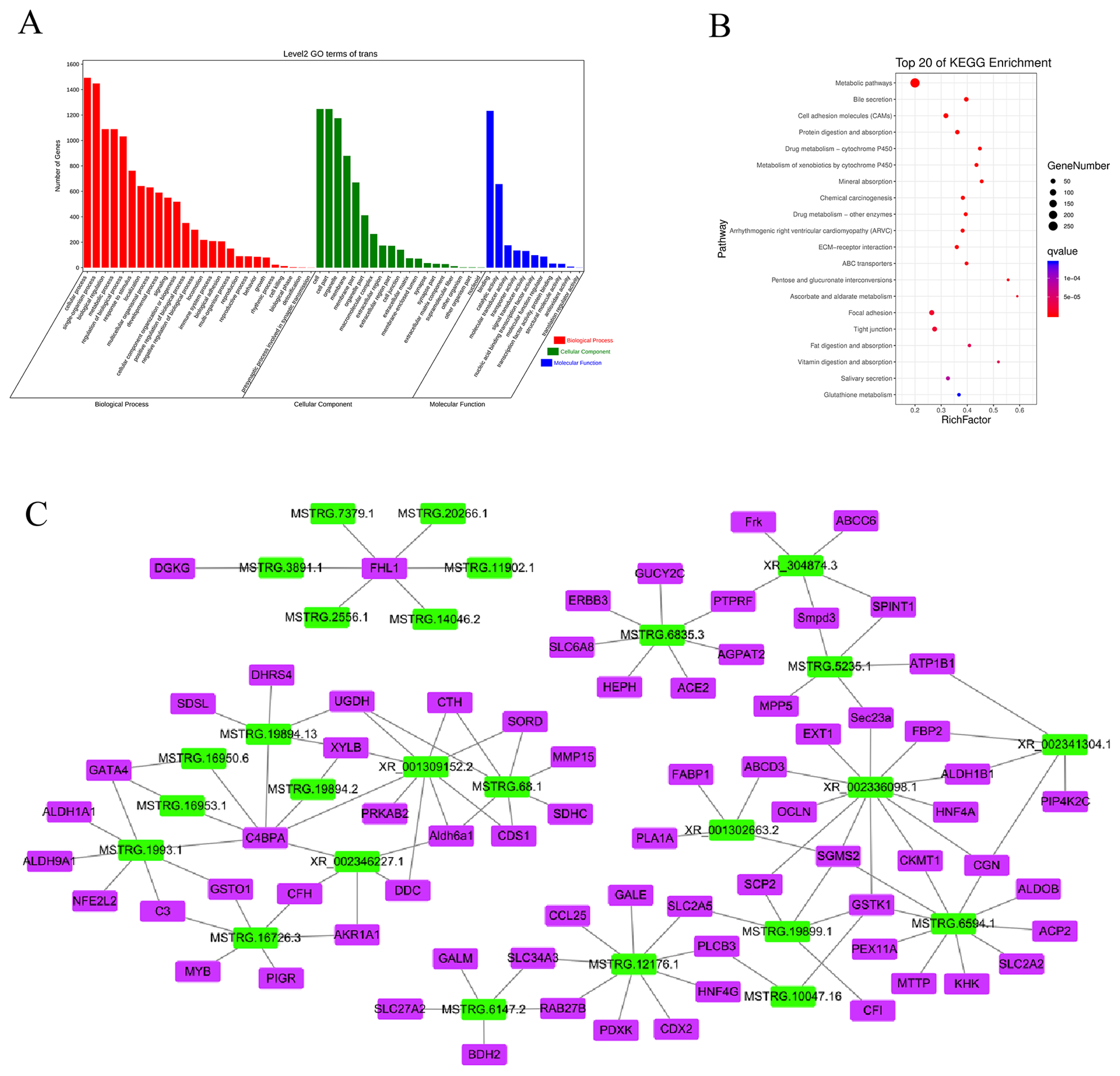
Figure 7
Accuracy of RT-qPCR verification results for the lncRNAs and mRNAs of Sus scrofa ileum under starvation stress. (A) RT-qPCR verification of lncRNAs. (B) RT-qPCR verification of mRNAs. a–c Indicates p<0.05, A–C indicates p<0.01.
Table 1
Base information before and after filtering and statistics of comparison analysis
REFERENCES
1. Blüher M. Obesity: global epidemiology and pathogenesis. Nat Rev Endocrinol 2019; 15:288–98.
https://doi.org/10.1038/s41574-019-0176-8


2. Yan T, Xiao R, Wang N, Shang R, Lin G. Obesity and severe coronavirus disease 2019: molecular mechanisms, paths forward, and therapeutic opportunities. Theranostics 2021; 11:8234–53.
https://doi.org/10.7150/thno.59293



3. Wardzinski E, Hyzy C, Duysen K, Melchert UH, Jauch-Chara K, Oltmanns KM. Hypocaloric dieting unsettles the neuroenergetic homeostasis in humans. Nutrients 2021; 13:3433
https://doi.org/10.3390/nu13103433



4. Varady K, Cienfuegos S, Ezpeleta M, Gabel K. Cardiometabolic benefits of intermittent fasting. Ann Rev Nutr 2021; 41:333–61.
https://doi.org/10.1146/annurev-nutr-052020-041327

5. Lee SR, Ko TH, Kim HK, et al. Influence of starvation on heart contractility and corticosterone level in rats. Pflugers Arch 2015; 467:2351–60.
https://doi.org/10.1007/s00424-015-1701-9


6. Martinez KB, Pierre JF, Chang EB. The gut microbiota: the gateway to improved metabolism. Gastroenterol Clin North Am 2016; 45:601–14.
https://doi.org/10.1016/j.gtc.2016.07.001



7. Christian VJ, Miller KR, Martindale RG. Food insecurity, malnutrition, and the microbiome. Curr Nutr Rep 2020; 9:356–60.
https://doi.org/10.1007/s13668-020-00342-0



8. Kazimierczyk M, Kasprowicz M, Kasprzyk M, Wrzesinski J. Human long noncoding RNA interactome: detection, characterization and function. Int J Mol Sci 2020; 21:1027
https://doi.org/10.3390/ijms21031027



9. Zhang X, Hong R, Chen W, Xu M, Wang L. The role of long noncoding RNA in major human disease. Bioorg Chem 2019; 92:103214
https://doi.org/10.1016/j.bioorg.2019.103214


10. Zou T, Jaladanki SK, Liu L, et al. H19 long noncoding RNA regulates intestinal epithelial barrier function via MicroRNA 675 by interacting with RNA-binding protein HuR. Mol Cell Biol 2016; 36:1332–41.
https://doi.org/10.1128/mcb.01030-15



11. Xiao L, Wu J, Wang JY, et al. Long noncoding RNA uc.173 promotes renewal of the intestinal mucosa by inducing degradation of microRNA 195. Gastroenterology 2018; 154:599–611.
https://doi.org/10.1053/j.gastro.2017.10.009



12. Wu F, Huang Y, Dong F, Kwon J. Ulcerative colitis-associated long noncoding RNA, BC012900, regulates intestinal epithelial cell apoptosis. Inflamm Bowel Dis 2016; 22:782–95.
https://doi.org/10.1097/mib.0000000000000691


13. Raychaudhuri S, Fan S, Kraus O, Shahinozzaman M, Obanda DN. Kale supplementation during high fat feeding improves metabolic health in a mouse model of obesity and insulin resistance. PloS one 2021; 16:e0256348
https://doi.org/10.1371/journal.pone.0256348



14. DeBry RW, Seldin MF. Human/mouse homology relationships. Genomics 1996; 33:337–51.
https://doi.org/10.1006/geno.1996.0209


15. Schook LB, Collares TV, Darfour-Oduro KA, et al. Unraveling the swine genome: implications for human health. Annu Rev Anim Biosci 2015; 3:219–44.
https://doi.org/10.1146/annurev-animal-022114-110815


16. Fouhse JM, Dawson K, Graugnard D, Dyck M, Willing BP. Dietary supplementation of weaned piglets with a yeast-derived mannan-rich fraction modulates cecal microbial profiles, jejunal morphology and gene expression. Animal 2019; 13:1591–8.
https://doi.org/10.1017/s1751731118003361


17. Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics (Oxford, England) 2018; 34:i884–i90.
https://doi.org/10.1093/bioinformatics/bty560



18. Langmead B, Salzberg S. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012; 9:357–9.
https://doi.org/10.1038/nmeth.1923



19. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102:15545–50.
https://doi.org/10.1073/pnas.0506580102



20. Cipriano A, Macino M, Buonaiuto G, et al. Wnt7bEpigenetic regulation of expression by the -acting long noncoding RNA Lnc-Rewind in muscle stem cells. eLife 2021; 10:e54782
https://doi.org/10.7554/eLife.54782



21. Cai B, Li Z, Ma M, et al. Six1LncRNA-Six1 encodes a micropeptide to activate in and is involved in cell proliferation and muscle growth. Front Physiol 2017; 8:230
https://doi.org/10.3389/fphys.2017.00230



22. Li J, Gao Z, Wang X, et al. Identification and functional analysis of long intergenic noncoding RNA genes in porcine pre-implantation embryonic development. Sci Rep 2016; 6:38333
https://doi.org/10.1038/srep38333



23. Jayakumar T, Chang CC, Lin SL, et al. Brazilin ameliorates high glucose-induced vascular inflammation via inhibiting ROS and CAMs production in human umbilical vein endothelial cells. Biomed Res Int 2014; 2014:403703
https://doi.org/10.1155/2014/403703



24. Phillips IR, Shephard EA. Drug metabolism by flavin-containing monooxygenases of human and mouse. Expert Opin Drug Metab Toxicol 2017; 13:167–81.
https://doi.org/10.1080/17425255.2017.1239718


25. Matsunaga T, Endo S, Maeda S, et al. Characterization of human DHRS4: an inducible short-chain dehydrogenase/reductase enzyme with 3beta-hydroxysteroid dehydrogenase activity. Arch Biochem Biophys 2008; 477:339–47.
https://doi.org/10.1016/j.abb.2008.06.002


26. Yamada T, Komoto J, Kasuya T, et al. A catalytic mechanism that explains a low catalytic activity of serine dehydratase like-1 from human cancer cells: crystal structure and site-directed mutagenesis studies. Biochim Biophys Acta 2008; 1780:809–18.
https://doi.org/10.1016/j.bbagen.2008.01.020


27. Spicer AP, Kaback LA, Smith TJ, Seldin MF. Molecular cloning and characterization of the human and mouse UDP-glucose dehydrogenase genes. J Biol Chem 1998; 273:25117–24.
https://doi.org/10.1074/jbc.273.39.25117


28. Zimmer BM, Barycki JJ, Simpson MA. Integration of sugar metabolism and proteoglycan synthesis by UDP-glucose dehydrogenase. J Histochem Cytochem 2021; 69:13–23.
https://doi.org/10.1369/0022155420947500



29. Guamán L, Oliveira-Filho E, Barba-Ostria C, et al. xylA and xylB overexpression as a successful strategy for improving xylose utilization and poly-3-hydroxybutyrate production in Burkholderia sacchari. J Ind Microbiol Biotechnol 2018; 45:165–73.
https://doi.org/10.1007/s10295-018-2007-7


30. Alzeer S, Ellis E. Metabolism of gamma hydroxybutyrate in human hepatoma HepG2 cells by the aldo-keto reductase AKR1A1. Biochem Pharmacol 2014; 92:499–505.
https://doi.org/10.1016/j.bcp.2014.09.004


31. Ursini F, Abenavoli L. The emerging role of complement C3 as a biomarker of insulin resistance and cardiometabolic diseases: preclinical and clinical evidence. Rev Recent Clin Trials 2018; 13:61–8.
https://doi.org/10.2174/1574887112666171128134552


32. García-Gen E, Penadés M, Mérida S, et al. High myopia and the complement system: factor h in myopic maculopathy. J Clin Med 2021; 10:2600
https://doi.org/10.3390/jcm10122600



33. Wang LK, Yue HL, Peng XJ, Zhang SJ. GSTO1 regards as a meritorious regulator in cutaneous malignant melanoma cells. Mol Cell Probes 2019; 48:101449
https://doi.org/10.1016/j.mcp.2019.101449


34. Cicirò Y, Sala A. MYB oncoproteins: emerging players and potential therapeutic targets in human cancer. Oncogenesis 2021; 10:19
https://doi.org/10.1038/s41389-021-00309-y



35. Zeng B, Wang H, Luo J, et al. Porcine milk-derived small extracellular vesicles promote intestinal immunoglobulin production through pIgR. Animals 2021; 11:1522
https://doi.org/10.3390/ani11061522



36. Xu B, Deng C, Wu X, et al. CCR9 and CCL25: A review of their roles in tumor promotion. J Cell Physiol 2020; 235:9121–32.
https://doi.org/10.1002/jcp.29782


37. Asgari-Karchekani S, Karimian M, Mazoochi T, Taheri M, Khamehchian T. CDX2 protein expression in colorectal cancer and its correlation with clinical and pathological characteristics, prognosis, and survival rate of patients. J Gastrointest Cancer 2020; 51:844–9.
https://doi.org/10.1007/s12029-019-00314-w


38. Daude N, Gallaher TK, Zeschnigk M, et al. Molecular cloning, characterization, and mapping of a full-length cDNA encoding human UDP-galactose 4’-epimerase. Biochem Mol Med 1995; 56:1–7.
https://doi.org/10.1006/bmme.1995.1048


39. He XX, Luo SS, Qin HQ, Mo XW. MicroRNA-766-3p-mediated downregulation of HNF4G inhibits proliferation in colorectal cancer cells through the PI3K/AKT pathway. Cancer Gene Ther 2021;
https://doi.org/10.1038/s41417-021-00362-0

40. Keller N, Mendoza-Ferreira N, Maroofian R, et al. Hereditary polyneuropathy with optic atrophy due to PDXK variant leading to impaired Vitamin B6 metabolism. Neuromuscul Disord 2020; 30:583–9.
https://doi.org/10.1016/j.nmd.2020.04.004


41. Rimessi A, Bezzerri V, Salvatori F, et al. PLCB3 loss of function reduces pseudomonas aeruginosa-dependent IL-8 release in cystic fibrosis. Am J Respir Cell Mol Biol 2018; 59:428–36.
https://doi.org/10.1165/rcmb.2017-0267OC


42. Jeon H, Kang SK, Lee MJ, et al. Rab27b regulates extracellular vesicle production in cells infected with Kaposi’s sarcoma-associated herpesvirus to promote cell survival and persistent infection. J Microbiol (Seoul, Korea) 2021; 59:522–9.
https://doi.org/10.1007/s12275-021-1108-6

43. Zwarts I, van Zutphen T, Kruit JK, et al. Identification of the fructose transporter GLUT5 (SLC2A5) as a novel target of nuclear receptor LXR. Sci Rep 2019; 9:9299
https://doi.org/10.1038/s41598-019-45803-x



44. Dhir G, Li D, Hakonarson H, Levine MA. Late-onset hereditary hypophosphatemic rickets with hypercalciuria (HHRH) due to mutation of SLC34A3/NPT2c. Bone 2017; 97:15–9.
https://doi.org/10.1016/j.bone.2016.12.001



- TOOLS






 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print





