 |
 |
| Anim Biosci > Volume 31(8); 2018 > Article |
|
Abstract
Objective
Present investigation was aimed to study the Single Nucleotide Variants of the luteinizing hormone beta (LHβ) gene and to analyze their association with the semen quality (fresh and post-thawed frozen semen) and luteinizing hormone (LH) concentrations in Murrah buffalo bulls.
Methods
Polymerase chain reaction–single stranded conformational polymorphism (PCR–SSCP) and Sanger sequencing method is used to study genetic variability in LHβ gene. LH assay was carried out using enzyme-linked immunosorbent assay method. A fixed general linear model was used to analyze association of single nucleotide polymorphism (SNP) of LHβ gene with semen quality in 109 and LH concentrations in 80 Murrah bulls.
Results
LHβ gene was found to be polymorphic. Total six SNPs were identified in LHβ gene g C356090A, g C356113T, g A356701G, g G355869A, g G356330C, and g G356606T. Single Stranded Conformational Polymorphism variants of pattern 2 of exon 1+pattern 2 of exon 2+pattern 1 of exon 3 had highly significant (p<0.01) effect on sperm concentration (million/mL), percent mass motility, acrosome integrity and membrane integrity in fresh and frozen semen whereas significant (p<0.05) effect was observed on percent live spermatozoa. SSCP variants of pattern 2 of exon 1+pattern 2 of exon 2+pattern 1 of exon 3 had highly significant (p<0.01) effect on luteinizing hormone concentrations too.
Selection of high quality bulls is a prerequisite for the cost-effective frozen semen production and dissemination of superior germplasm. However, direct selection for semen quality traits is not possible because of their low heritability [1]. Therefore, marker assisted selection could be used for selection of superior males at an early age. With the development of molecular biological techniques, the candidate gene method might offer the specific markers to predict sperm quality traits in bulls. Researchers [2] indicated that hormone and hormone receptors are good candidate genes for bull sperm traits due to their modulating roles in many male reproductive pathways, thus they are considered to be good candidate genes for the reproductive traits.
Luteinizing hormone (LH) is a pituitary-derived heterodimeric glycoprotein, consisting of the common α-subunit with human chorionic gonadotropin, follicle stimulating hormone and thyroid stimulating hormone and a unique β-subunit conferring the biological specificity of LH [3,4]. Luteinizing hormone beta polypeptide (LHβ) gene is a single copy gene [5], located on the long (q) arm of chromosome 18 and consists of 3 exons. Studies indicated that mutation in LH beta subunit, causes its inactivation leading to lack of leydig cells, absence of spontaneous puberty and infertility in human [6] and a genetic variant of the LHβ might affect susceptibility to prostate cancer via altered testosterone secretion [7]. The polymorphism study of LHβ gene has been done [8], however, a study regarding the use of this gene as candidate gene marker for sperm quality traits and hormone concentrations in Murrah bulls is yet to be conducted. Therefore, present investigation was undertaken to study the single nucleotide variants of the LHβ gene and to analyze their association with the semen quality (fresh and post-thawed frozen semen) and LH concentrations in Murrah bulls.
All the experimental procedures were conducted as per the guidelines of the Institutes Animal Ethics Committee (IAEC) of ICAR-NDRI, Bengaluru, India.
The present investigation was carried out in Murrah bulls maintained at State Livestock Breeding and Training Center (SLBTC) Hessarghatta, Bengaluru, Nandini Sperm Station (NSS), Hessarghatta, Bengaluru, and Centralized Semen Collection Centre of Livestock Breeding and Training Centre (LBTC), Dharwad. Bulls were maintained under standard management conditions adhering to Minimum Standard Protocols as recommended by the Department of Dairying, Animal Husbandry & Fisheries, Government of India.
About ten milliliters of blood was collected aseptically from jugular vein of each animal (n = 109) in a vacutainer tube containing 0.5% ethylenediaminetetraacetic acid (EDTA). After collection, the samples were stored at 4°C and DNA was isolated within 24 hours of collection by high salt method [9]. The working solution of DNA was prepared by diluting the stock to 100 ng/μL for utilizing as DNA template in polymerase chain reaction (PCR).
For LH level estimation in bulls (n = 80), five milliliters blood was collected in serum separator tube (SST) immediately after ejaculation because as per [10] sexual stimulation such as site of a cow, teasing and on one occasion the act of ejaculation caused an immediate release of large amount of LH. Blood samples were allowed to clot and centrifuged at 1,000×g for 15 minutes, from which serum samples were aliquoted and stored at −20°C to −80°C until analyzed for LH concentrations.
Serum LH concentrations were determined by CUSABIO Bovine LH ELISA Kit. Calculations were done using the professional software “cure expert 1.3” developed by Hyams. Duplicate readings for each standard and sample were averaged and optical density of blank was subtracted from standard and sample. A standard curve was established by reducing the data using computer software capable of generating four parametric logistic (4-PL) curve fit and LH concentrations was calculated. Sensitivity was less than 1.56 mIU/mL and intra- and inter-assay coefficients of variation were <15%.
Semen quality traits like semen volume per ejaculate, sperm concentration (million/mL), percent individual motility, percent non-eosinophilic spermatozoa, percent normal spermatozoa, percent membrane integrity in both fresh and post-thawed frozen semen were obtained from each ejaculate according to the guidelines of the World Health Organization [11]. The semen samples were collected from each bull in 3 to 6 days intervals using artificial vagina during three different seasons viz., rainy (July to October), winter (November to February) and summer (March to June). Two ejaculates per bull in a season were used for semen quality evaluation and mean value obtained was used for association studies.
Immediately after collection, the ejaculates were stored at 37°C in water bath prior to the evaluation for semen quality traits. The ejaculate volume was measured in a sperm collecting vial and counted. Sperm concentration (million/mL) was determined by using a digital photometer with auto dilutor (SMILE software, IMV Technologies, Paris, France). Percent live spermatozoa, percent membrane integrity, percent acrosome integrity and percent normal spermatozoa were determined using standard protocols. The percent live spermatozoa were estimated by Eosin-Nigrosin staining [12], wherein the dead cells take the eosin stain, whereas live cells remain un-stained. Percent membrane integrity of spermatozoa was determined by using HOST. The percentage of hypo-osmotic swelling test reacted cells (cells with coiled tail) in hypo osmotic solution (150 mOsmol/L) was determined [13]. Percent acrosomal integrity of the spermatozoa was studied by Giemsa’s staining of fixed smears and cells with intact acrosome were counted [14]. After storage in liquid nitrogen for 5 to 7 days, two straws were randomly obtained from each bull, thawed at 37°C for 30 s, and immediately evaluated for the percent post thaw sperm motility according to the guidelines of the World Health Organization [11]. In brief, the post thaw spermatozoa were viewed on a TV monitor connected to a camera mounted onto a phase contrast microscope (Olympus-BX40, Minitub, Tiefenbach, Germany) at 100× magnification by placing a drop of semen on to a pre-warmed (37°C) slide and overlaying it with a slip cover.
Three sets of overlapping primers were designed for LHβ gene based on reference sequence of Bubalus bubalis (NCBI Reference Sequence: NW_005783642.1) by using primer 3 (V.0.4.0) (http://primer3.ut.ee/) online software for amplifying the complete gene and were procured from Sigma-Aldrich Chemicals Pvt. Ltd. Bengaluru, India. The details of the primers, targeted region, annealing temperature, and expected product sizes are summarized in Table 1.
The PCR was carried out on approximately 100 ng of genomic DNA in 25 μL per reaction volume. The PCR reaction mixture consisted of 200 μM of each dNTPs, 10× Taq Pol assay buffer, 1 U Taq polymerase enzyme (Genet Bio, Daejeon, Korea) and 20 pM of each primer (Sigma Aldrich, St. Louis, MO, USA). The thermocycler conditions included an initial denaturation at 94°C for 2 min, followed by 35 cycles of denaturation at 94°C for 30 s with varying annealing temperatures based on primer set (Table 1), extension at 72°C for 1 min, followed by a final extension at 72°C for 10 min. The PCR products were electrophoresed at 100 V in 1.5% agarose gel in 1×tris borate EDTA (TBE) (Sigma Aldrich, USA) buffer containing 0.5 μg/mL ethidium bromide (Sigma Aldrich, USA) along with a DNA molecular size marker of 100 bp (Sigma Aldrich, USA). The gels were visualized and documented using Gel documentation system (Gel doc 1000, Bio-Rad, Hercules, CA, USA).
The genetic variants were determined by single strand conformation polymorphism (SSCP) technique. Amplified PCR products (10 μL) were further diluted in 10 μL of denaturing solution (95% formamide, 10 mM NaOH, 0.05% xylene cyanol, 0.05% bromophenol blue, and 20 mM EDTA, Sigma Aldrich, USA) and denaturation was carried out at 95°C for 5 min followed by rapid chilling on an ice block for 20 min and loaded on 10% acrylamide:bisacrylamide (29:1, Sigma Aldrich, USA) in 1× TBE (Sigma Aldrich, USA) buffer for 15 h (200 V) at 4°C. The gels were silver stained as described by Sambrook and Russell [15]. Band patterns were characterized by the number of bands and mobility shifts, and each pattern was scored manually. To confirm the mobility shift in each pattern, PCR products of each SSCP pattern in duplicates were chosen and custom sequenced using automated ABI DNA Sequencer 3730 XL (Thermo Fisher Scientific, Meridian Road, Rockford, IL, USA) for detecting single nucleotide polymorphisms (SNPs). Sequence data were analyzed using DNA Baser (Heracle BioSoft SRL, Pitesti, Romania) and Clustal W multiple sequence alignment software (developed by Des Higgins) for detecting SNPs [16] by comparing the observed sequence of the LHβ gene in Murrah bulls with the NCBI Resource Coordinators (2016). Database resources of the National Center for Biotechnology Information. Nucleic Acids Research, 44(Database issue), D7–D19. http://doi.org/10.1093/nar/gkv1290) reference sequence for Bubalus bubalis.
The associations between SSCP pattern set of three exons in LHβ gene and semen quality traits were analyzed using the general linear model (GLM) and compared by Duncan’s multiple range test of SPSS software (Version 16.0, SPSS Inc. Released 2007. SPSS for Windows, Chicago, IL, USA).
The linear model is represented as follows:
Where, Yijklm = observed value of mth bull in ith herd, jth age group, kth season and lth genotype for each semen quality trait. μ = overall mean, Ii = fixed effect of ith herd (1 = State Livestock Breeding and Training Centre, 2 = Nandini Sperm Station and 3 = Centralized Semen Collection Centre of Livestock Breeding and Training Centre), Aj = fixed effect of jth age (1 = 2–4 years, 2 = 4–6 years, 3 = 6–8 years and 4 = ≥8 years), Sk = fixed effect of kth season (1 = rainy, 2 = winter, and 3 = summer), Gl = fixed effect of Ith SSCP pattern set of three exons, eijklm = random error which is normally and independently distributed with mean 0 and variance σ2e).
The association between LHβ SSCP pattern set and LH concentrations was studied using the GLM and compared by Duncan’s multiple range test of SPSS software (Version 16.0, SPSS Inc. Released 2007. SPSS for Windows, USA). The linear model is represented as follows:
Where, Yij = observation of jth bull having ith genotype for hormone concentration. μ = overall mean, Gi = fixed effect of ith SSCP pattern set, eij = random error is normally and independently distributed with mean 0 and variance σ2e.
The polymorphism in LHβ gene was investigated using three sets of overlapping primers by PCR-SSCP analysis. Exon 1 showed polymorphism within the population. Unique SSCP patterns with different mobility shifts, pattern 1 and pattern 2 were observed (Figure 1). The frequency of pattern 1 and pattern 2 were 0.2018 and 0.7982, respectively. PCR-SSCP analysis of exon 2 showed polymorphism within the population. Unique SSCP patterns with different mobility shifts, pattern 1, pattern 2, and pattern 3 were observed (Figure 2). The frequency of SSCP variants pattern 1, pattern 2, and pattern 3 were 0.2844, 0.5046, and 0.2110, respectively and similarly, PCR-SSCP analysis of exon 3 showed two unique SSCP patterns (Figure 3). Pattern 1 and pattern 2 with frequency of 0.4954 and 0.5046, respectively. SSCP band patterns along with their frequency are shown in Table 2.
PCR products showing unique SSCP patterns were custom sequenced and analyzed. The analysis revealed one silent mutation g G355869A in the partial intron 1 region (Table 3). Pattern 1 was similar to the Bubalus bubalis reference sequence having G at position 355869 whereas pattern 2 bulls were heterozygous with A/G at this site.
In exon 2, two transversions, g C356090A and g G356330C and one transition g C356113T were observed. Out of these, C356090A and g C356113T were present in the coding region while g G356330C was present in the intron 2. At position 356090, pattern 1, pattern 2, and pattern 3 were carrying C, A/C, and C nucleotides, respectively. At position 356113, pattern 1, pattern 2, and pattern 3 were carrying C, C, and T/C nucleotides, respectively. At position 356330, pattern 1, pattern 2, and pattern 3 were carrying G, C/G, and G nucleotides, respectively. Pattern 1 was similar to the reference Bubalus bubalis sequence whereas pattern 2 was different from pattern 1. All mutations found in the LHβ exon 2 were silent mutations (Table 3).
In the present study, exon 3 was found to be polymorphic at two sites leading to transition at position g A356701G in the coding region and transversion at g G356606T in the third intron of LHβ gene as compared to Bubalus bubalis reference sequence. Mutation at g A356701G leads to change in amino acid from Histidine to Arginine (Table 3). The bulls showing pattern 1 were homozygous at the two sites with A and G nucleotides respectively and were similar to reference sequence. The bulls with pattern 2 were heterozygous at both sites with G/A and T/G respectively. Chromatograph analysis of first, second and third fragments of LHβ gene is shown in Figures 4, 5, 6, respectively. Sequence for LHβ gene was submitted to Gene Bank with the accession number KY786093. SNP’s identified in the LHβ gene of Murrah bulls were compared with other bovine species having transcript ENSBTAT00000057054.1. None of the SNP’s identified in the LHβ gene of Murrah bulls was similar to other bovines, thereby reflecting their specificity to LHβ gene of Murrah bulls.
SSCP variants of pattern 2 of exon 1+pattern 2 of exon 2+pattern 1 of exon 3 had highly significant effect (p<0.01) on sperm concentration (million/mL), percent mass motility, acrosome integrity and membrane integrity in fresh and frozen semen whereas significant (p<0.05) effect was observed on percent live spermatozoa. Bulls with pattern 2 of exon 1+pattern 2 of exon 2+pattern 1 of exon 3 had the highest sperm concentration (1,334.11±74.04×106/mL), percent mass motility (71.85± 1.32), percent live spermatozoa (74.85±2.17) fresh semen percent acrosome integrity (77.20±1.20), fresh semen percent membrane integrity (72.75±1.41), frozen semen percent acrosome integrity (70.30±1.32) and frozen semen percent membrane integrity (66.02±0.95) compared to bulls with pattern 2 of exon 1 +pattern 1 of exon 2+pattern 2 of exon 3 and pattern 2 of exon 1+pattern 2 of exon 2+pattern 2 of exon 3 (Table 4).
SSCP variants of pattern 2 of exon 1+pattern 2 of exon 2+pattern 1 of exon 3 had highly significant (p<0.01) effect on LH concentrations. Bulls with pattern 2 of exon 1+pattern 2 of exon 2+pattern 1 of exon 3 had the highest LH concentrations (26.98±2.58) compared to pattern 2 of exon 1+pattern 2 of exon 2+pattern 2 of exon 3 (19.23±2.94) (Table 5).
PCR-SSCP is a simple, sensitive and efficient technique for identifying the genetic variations in candidate genes. The study revealed six SNPs in the LHβ gene in Murrah bulls. Our findings regarding characterization of LHβ gene were in agreement with the findings of earlier researchers [13]. They reported seven different SNPs in the LHβ gene in Indian river buffalo by using PCR-SSCP technique and direct sequencing methods. In all variants, amino acid substitutions were noted. Therefore, LHβ gene was not highly conserved and non-synonymous mutations were observed. In comprehensive view, it appears that the bulls with pattern 2 of exon 1+pattern 2 of exon 2+pattern 1 of exon 3 could be used as a marker for selecting higher sperm concentration, mass motility, live spermatozoa acrosome integrity and membrane integrity. In the present study, coding as well as non-coding regions of LHβ gene was highly polymorphic and had strong association with semen quality parameters in Murrah bulls. Our findings are in agreement with those of the previous investigation [17]. They also reported the significant association of LHβ gene polymorphism with semen concentration in Chinese Buffaloes.
The hormones and their receptors genes are good candidate markers for bull semen traits due to their modulating roles in many male reproductive pathways [2]. Our results indicated that genetic polymorphism in LHβ gene lead to variation in LH concentrations which could directly affect the quality of semen. The Murrah bulls with pattern 2 of exon 1+pattern 2 of exon 2+pattern 1 of exon 3 could be selected as a marker for higher LH concentrations because these bulls had higher semen concentration, percent mass motility, percent live spermatozoa, percent acrosome integrity and membrane integrity. Published reports are not available to compare the association of SSCP variants of LHβ gene with LH concentrations in Murrah bulls. Therefore, findings of this study are compared with those reported in cattle. Some researchers [18] suggested that the mutations in the 5′ upstream regulation regions would have altered the binding sites of transcription factors affecting the transcription leading to variation in FSH concentrations in cattle. Consequently, it becomes very important to identify mutations and their impact on traits of interest because all mutations might not be favorable and some can interrupt cellular functions thereby decreasing the performance of important economic traits.
Based on the findings from the present investigation, it is concluded that LHβ gene exhibited high degree of genetic variability in Murrah bulls. The identified SSCP pattern set with pattern 2 of exon 1+pattern 2 of exon 2+pattern 1 of exon 3 is significantly associated with semen quality parameters and hormone concentrations, which could be used as a genetic marker in Marker Assisted Selection after validating the same SSCP pattern using large number of Murrah bulls from different herds.
ACKNOWLEDGMENTS
The Authors are thankful to the Director, NDRI, Karnal and Head, SRS of NDRI, Bengaluru for providing the necessary facilities, financial assistance and also to the Director of “Karnataka Livestock Development Agency” (KLDA), Government of Karnataka, for providing necessary facilities to work in their organized semen stations.
Figure 1
Polymerase chain reaction–single stranded conformational polymorphism pattern of exon 1 of luteinizing hormone beta gene.

Figure 2
Polymerase chain reaction–single stranded conformational polymorphism pattern of exon 2 of luteinizing hormone beta gene.

Figure 3
Polymerase chain reaction–single stranded conformational polymorphism pattern of exon 3 of luteinizing hormone beta gene.
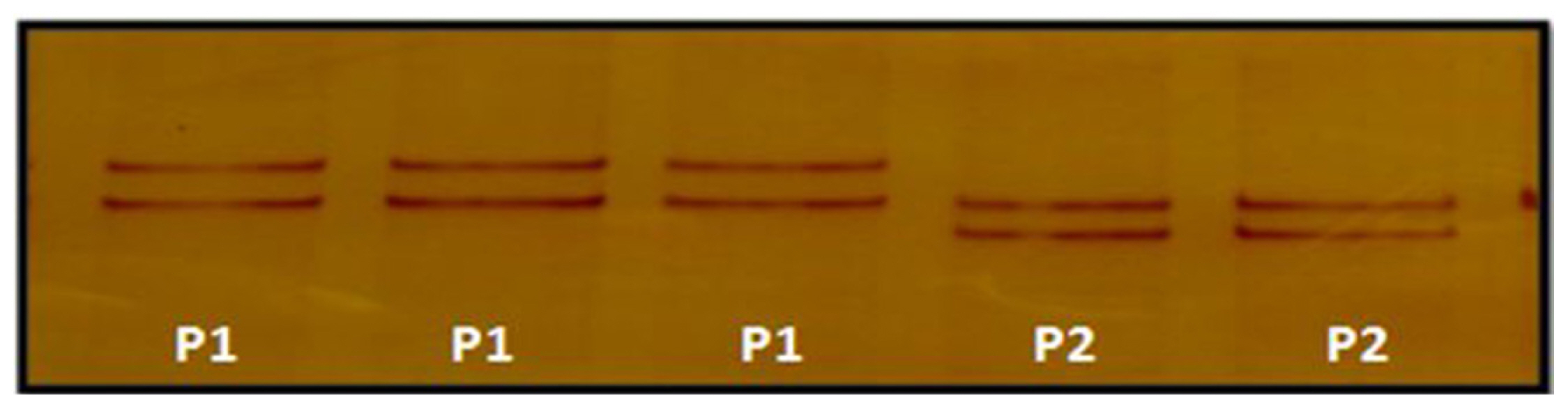
Figure 4
Sanger traces figures of single stranded conformational polymorphism variant sites of exon 1 of luteinizing hormone beta gene.

Figure 5
Sanger traces figures of single stranded conformational polymorphism variant sites of exon 2 of luteinizing hormone beta gene.
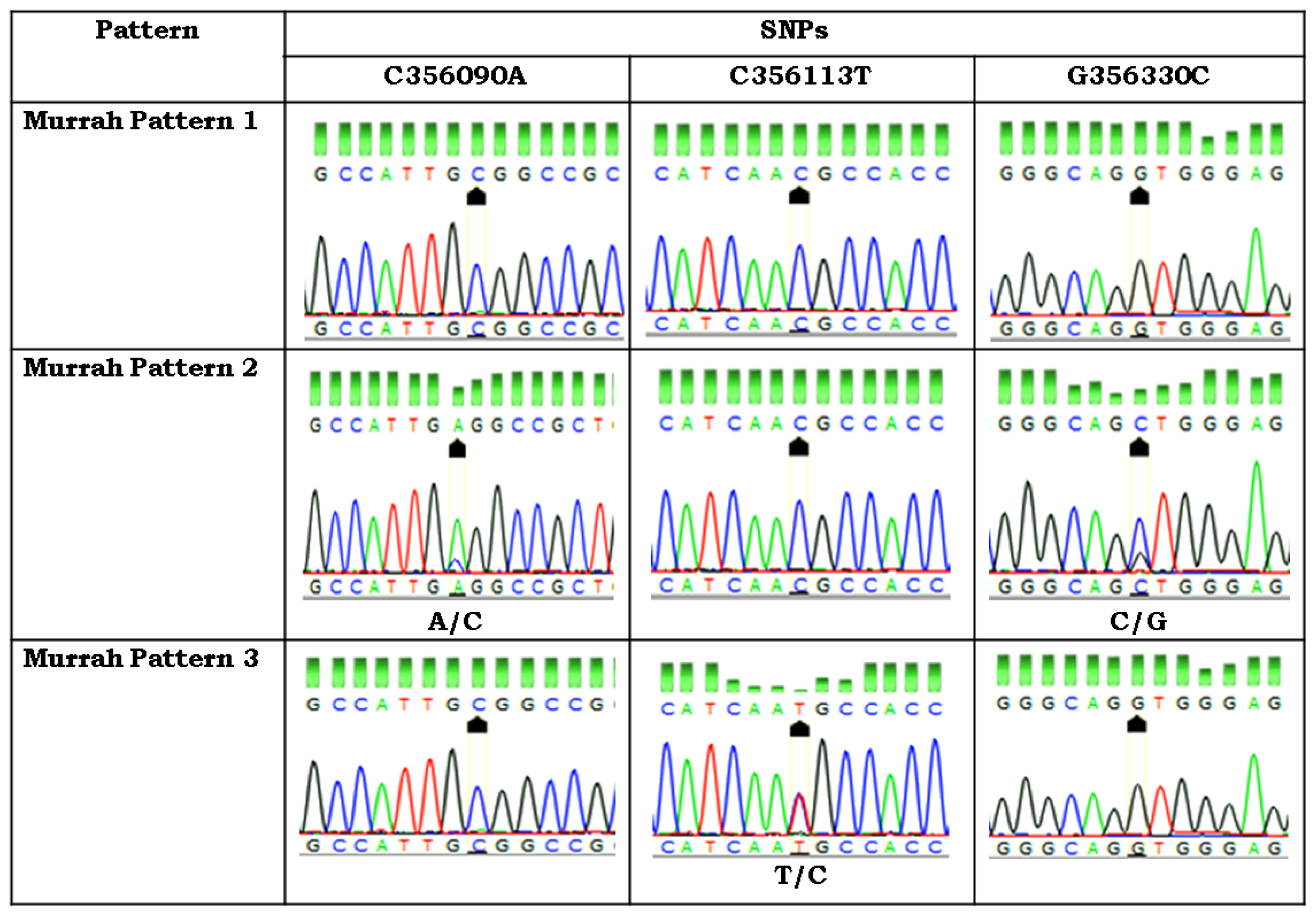
Figure 6
Sanger traces figures of single stranded conformational polymorphism variant sites of exon 3 of luteinizing hormone beta gene.

Table 1
Primer sequences (5′ to 3′) used for amplification of LHβ gene
Table 2
Frequency of SSCP variants of LHβ gene
Table 3
Summary of single nucleotide polymorphism observed in LHβ gene in Murrah bulls
Table 4
Effect of combined patterns of three exons of the LHβ gene on the semen quality parameters (mean±SE)
| Effects1) | Concentration (millions of cells/mL) | Volume (mL) | Mass motility (%) | Post thaw motility (%) | Live (%) | Normal (%) | Head (%) | Mid piece (%) | Tail (%) | Fresh Giemsa (%) | Fresh HOST (%) | Frozen Giemsa (%) | Frozen HOST (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||||||
| Abnormalities | |||||||||||||
| P2E1+P2E2+P1E3 | 1,334.11±74.04b | 5.80±0.24 | 71.85±1.32a | 45.70±0.30 | 74.85±2.17a | 88.97±0.91 | 7.50±0.20 | 2.70±0.32 | 6.02±0.63 | 77.20±1.20a | 72.75±1.41a | 70.30±1.32a | 66.02±0.95a |
| P2E1+P1E2+P2E3 | 961.66±124.63a | 4.58±0.41 | 65.41±1.12b | 43.00±0.51 | 70.41±3.66b | 89.41±1.53 | 7.33±0.33 | 3.41±0.55 | 5.33±1.06 | 70.89±1.61b | 68.76±1.50b | 68.23±1.77ab | 63.25±1.60ab |
| P2E1+P2E2+P2E3 | 1,309.11±101.76b | 4.66±0.33 | 69.05±1.82a | 44.46±0.41 | 69.23±2.99b | 86.77±1.25 | 7.11±0.27 | 3.44±0.45 | 6.33±0.87 | 73.41±0.97ab | 67.53±1.70b | 66.82±0.98b | 60.71±1.31b |
| p-value | 0.002* | 0.877 | 0.001** | 0.701 | 0.006* | 0.294 | 0.360 | 0.324 | 0.307 | 0.001** | 0.001** | 0.001** | 0.001** |
1) P2E1, pattern 2 of exon 1; P2E2, pattern 2 of exon 2; P1E3, pattern 1 of exon 3; P1E2, pattern 1 of exon 2; P2E3, pattern 2 of exon 3.
Table 5
Effect of combined patterns of three exons of the LHβ gene on Luteinizing hormone concentration (mean±SE)
| Effects1) | Luteinizing hormone concentration (mIU/mL) |
|---|---|
| P2E1+P2E2+P1E3 | 26.98±2.58a |
| P2E1+P2E2+P2E3 | 19.23±2.99b |
| p-value | 0.001** |
REFERENCES
1. Mathevon M, Buhr MM, Dekkers JCM. Environmental, management and genetic factors affecting semen production in Holstein bulls. J Dairy Sci 1998; 81:3321–30.


2. Giesecke K, Hamann H, Sieme H, Distl O. INHBA-associated markers as candidates for stallion fertility. Reprod Domest Anim 2010; 45:34247

3. Pierce JG, Parsons TF. Glycoprotein hormones structure and function. Ann Rev Biochem 1981; 50:465–95.


4. Gharib SD, Wierman ME, Shupnik MA, Chin WW. Molecular biology of the pituitary gonadotropins. Endocr Rev 1990; 11:177–99.



5. Talmadge K, Booratein WR, Fiddes JC. The human genome contains seven genes for the β-subunit of chorionic gonadotropin but only one gene for the β-subunit of luteinizing hormone. DNA 2009; 2:281–9.

6. Huhtaniemi I, Jiang M, Nilsson C, Pettersson K. Mutations and polymorphisms in gonadotropin genes. Mol Cell Endocrinol 1999; 25:89–94.

7. Elkins DA, Yokomizo A, Thibodeau SN. Luteinizing hormone beta polymorphism and risk of familial and sporadic prostate cancer. Prostate 2003; 56:30–6.


8. Basavarajappa MS, De S, Thakur M, et al. Characterization of the luteinizing hormone beta (LHβ) subunit gene in the Indian River buffalo (Bubalus bubalis). Gen Comp Endocrinol 2008; 155:63–9.


9. Miller SA, Dykes D, Polesky HF. A sample salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988; 16:1215




10. Katongole CB, Naftolin F, Short RV. Relationship between blood levels of luteinizing hormone and testosterone in bulls, and the effects of sexual stimulation. J Endocrinol 1971; 50:457–66.


11. World Health Organization. WHO laboratory manual for the examination of human semen and semen-cervical mucus interaction. 4th edNew York, USA: Cambridge University Press; 1999.
12. Blom EA. Rapid staining method using eosin-nigrosin to distinguish between live and dead spermatozoa. Nord Vet Med 1950; 18:1390
13. Muhammad Z, Akbar LL, Ahmad E, Muhammad G. Hypo osmotic swelling test as screening for evaluation of semen of bull. J Entomol Zool Stud 2013; 1:124–8.
14. Correa JR, Zavos PM. The hypo-osmotic swelling test; its employment as an assay to evaluate the functional integrity of the frozen-thawed bovine sperm membrane. Theriogenology 1994; 42:351–60.


15. Sambrook J, Russell DW. Molecular cloning. A laboratory manual. 3rd edCold Spring Harbor, NY, USA: Cold Spring Laboratory Press; 2001.
16. Hall TA. Bio-Edit: a user-friendly biological sequence alignment editor analysis program for Windows 95/98/ NT. Nucleic Acids Symp Ser 1999; 41:95–8.
- TOOLS






 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print





