 |
 |
| Anim Biosci > Volume 31(4); 2018 > Article |
|
Abstract
Objective
This study was conducted to isolate the cellulolytic microorganism from the rumen of Holstein steers and characterize endoglucanase gene (Cel5A) from the isolated microorganism.
Methods
To isolate anaerobic microbes having endoglucanase, rumen fluid was obtained from Holstein steers fed roughage diet. The isolated anaerobic bacteria had 98% similarity with Eubacterium cellulosolvens (E. cellulosolvens) Ce2 (Accession number: AB163733). The Cel5A from isolated E. cellulolsovens sp. was cloned using the published genome sequence and expressed through the Escherichia coli BL21.
Results
The maximum activity of recombinant Cel5A (rCel5A) was observed at 50°C and pH 4.0. The enzyme was constant at the temperature range of 20°C to 40°C but also, at the pH range of 3 to 9. The metal ions including Ca2+, K+, Ni2+, Mg2+, and Fe2+ increased the endoglucanase activity but the addition of Mn2+, Cu2+, and Zn2+ decreased. The Km and Vmax value of rCel5A were 14.05 mg/mL and 45.66 μmol/min/mg. Turnover number, Kcat and catalytic efficiency, Kcat/Km values of rCel5A was 96.69 (s−1) and 6.88 (mL/mg/s), respectively.
Cellulose is one of the most abundant renewable organic compound in the environment, about 100 billion dry tons per year is produced in the biosphere through photosynthesis [1], and is considered as the most important waste material produced in the agriculture industry [2]. Cellulolytic microorganisms can degrade cellulosic materials using the multiple enzyme components which are classified as the cellulase system. There are three major types of enzymes in cellulase system: [β-1,4-endoglucanase (endocellulases, EC 3.2.1.4), which randomly hydrolyze internal bonds), cellobiohydrolase (exocellulases, EC 3.2.1.91), cleave the ends of the cellulose chains results in cellotetraose or cellobiose) and β-glucosidases (EC 3.2.1.21), which produce monosaccharide, glucose from cellobiose [3]. These classified enzymes can be fractionated by its characteristics which have function to contribute to the cellulase system.
Ruminants are species possessing the most powerful ability to utilize of plant cell wall content because of abundant rumen microorganisms which can degrade plants [4]. The enzymes which have plant cell-wall degrading activity are some of the major and most studied systems in the rumen microbial ecology [5]. Degradation of cell wall content is related to the complex of ruminal cellulolytic microorganisms [6]. Ruminal fungi have the role of producing fibrolytic enzymes and utilizing the recalcitrant fiber fraction [7]. There is a previous study indicating that protozoa also have some ability of fiber digestion [8]. Although, the major fibrolytic ruminal bacteria are Fibrobacter succinogenes, Ruminococcus albus, and Ruminococcus flavefaciens [9], the genus Eubacterium containing many species with a wide spectrum of phenotypes, has been discovered in the rumen [10]. Among them, the fibrolytic species Eubacterium cellulosolvens (E. cellulosolvens) can dominate especially when selective substrate such as cellulose is used [10]. In the case of European cattle, E. cellulosolvens is one of the predominant cellulolytic bacteria in the rumen [11].
To our knowledge, a study dealing with the detailed characterization of cellulolytic activity of E. cellulosolvens has not been extensively conducted. Accordingly, the endoglucanase gene (Cel5A) was cloned from E. cellulosolvens isolated from Holstein steers and the recombinant enzyme was characterized in detail to prove the usability of the gene’s enzymatic value for further application in this study.
This study used two rumen fistulated Holstein steers managed at the Center for Animal Science Research, Seoul National University, Korea. Animal use and protocols were reviewed and approved by the Seoul National University Animal Research Ethics Committee.
The rumen fluid was obtained from the rumen-fistulated Holstein steers fed alfalfa and commercial concentrates and kept anaerobic condition using stainless steel vacuum insulated bottle and directly transferred to the laboratory. The rumen fluid was filtered through 8 layers of cheesecloth and homogenized using a mixer for 1 min. To maintain anaerobic condition, flushing the oxygen-free CO2 gas into the bottle was done continuously. Then, the prepared rumen fluid was inoculated into sterilized Dehority’s artificial medium [12] in the Hungate tubes and serially diluted 104 to 106 fold with same medium. The dilutions from each tubes were spread (100 μL) on the plate having chromogenic carboxymethylcellulose (Azo-CMC, Megazyme Inc., Dublin, Ireland) and incubated at 39°C for 48 h in anaerobic chamber filled with 95% of CO2 and 5% of H2.
After incubation, the cellulolytic microorganisms were screened through the presence of yellow halos around the colonies (Figure 1). A total of 18 colonies were screened and inoculated into the fresh Dehority liquid medium, and incubated for 3 days. The grown microorganisms were subcultured twice as described above. For identification of the selected bacterial strain, 16S ribosomal RNA gene sequence was amplified by polymerase chain reaction (PCR). Bacterial cell pellet was obtained from each isolated culture by centrifugation (10,000 g×1 min) prior to DNA extraction, which was performed with aid of DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany). The universal primers 27f (5′-AG AGTTTGATCMTGGCTCAG-3′) and 1492r (5′-ACGGCTACC TTGTTACGACTT-3′) were used to amplify the 16S rRNA gene fragments using PCR [13]. The amplification was carried out using HotStar Taq Master Mix Kit (Qiagen, Germany) with the following cycling parameter: 95°C for 15 min, followed by 35 cycles of 30 s at 94°C, 30 s at 55°C, and 90 s at 72°C with final extension for 7 min. The amplified PCR product was loaded on agarose gel. The 16S rRNA band was purified using a Gel DNA extraction kit (Qiagen, Germany). A sequence similarity search of the purified PCR product was done using BLAST in National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov).
Escherichia coli (E. coli) BL21 (DE3) (Novagen pET Systems, Merck Millipore Corp., Darmstadt, Germany) were used for the constructed vector’s host. The E. coli strain was grown in 37°C in Luria-Bertani (LB) Broth containing 30 μg/mL kanamycin with shaking (180 rpm) and the isolated bacteria (E. cellulosolvens sp.) were incubated anaerobically in 39°C in Dehority’s artificial medium containing 1% of carboxymethylcellulose (CMC) without agitation.
Genomic DNA and plasmid DNA were purified using G spin TM Genomic DNA extraction kit and plasmid DNA extraction kit (iNtRON Inc., Seoul, Korea) respectively. The plasmid vector, pET41b (Novagen’s pET Systems, Merck Millipore Corp., Germany) is used for cloning and expression of Cel5A. For amplification of the Cel5A gene’s open reading frame (ORF) from genomic DNA of E. cellulosolvens, AccuPower PCR PreMix (Bioneer Inc., Daejeon, Korea) and primer set EC-f (5′-AG ATATACATATGAAAGGAAACTGGTTG-3′ with NdeI site) and EC-r (5′-AGATATACTCGAGCGGGATCAGGTTGC-3′ with XhoI site) were used and PCR was performed with the following cycling temperature and time: 95°C for 15 min, followed by 40 cycles of 30 s at 94°C, 1 min at 60°C, and 3 min 30 s at 72°C with final elongation for 10 min. The DNA fragment of Cel5A and pET-41b was digested with the same restriction enzymes (NdeI and XhoI). The enzyme treated target DNA and vector was purified from the Agarose gel and ligated, then transformed into E. coli T10F′ for checking its sequence using NCBI sequence database. The recombinant plasmid was purified from E. coli T10F′ and then transformed into E. coli BL21 (DE3) for protein expression. The transformant was identified by colony PCR [14] using same primer set and PCR condition. The transformants were incubated overnight in LB media containing kanamycin (30 μg/mL) and the culture was used as the DNA templates for the colony PCR. The PCR products were electrophoresed with 0.8% agarose gel. The isolates which had the clear 3.5 kb band at the gel, were selected for the further experiment.
A total of 150 mL of LB broth containing 30 μg/mL kanamycin was prepared and inoculated with transformed E. coli with pET41b-Cel5A then grown at 37°C for 3 h in a shaking incubator set at 180 rpm. After the optical density in 600 nm (OD600) value of the culture reached around 1.0, Isopropyl β-D-1-thiogalactopyranoside (IPTG) 1 mM was added then shook overnight in 16°C. The cell pellet was collected after centrifugation at 10,000 g for 10 min and then lysed by Bugbuster reagent (Novagen’s pET Systems, Merck Millipore Corp., Germany). The cell lysate was harvested by centrifugation at 12,000 g for 20 min and the His-tagged Cel5A was purified using Ni-NTA His-Bind column (Novagen’s pET Systems, Merck Millipore Corp., Germany). A gradient of 5 to 250 mM imidazole in Tris-HCl buffer containing 500 mM NaCl was employed to purify the fusion protein. After elution of the target protein from the resin, the product was used for further characterization of its properties.
Endoglucanase activity was determined using 3, 5-dinitrosalicylic acid (DNS) reagent [15]. The purified enzyme with the addition of 1% (w/v) of CMC prepared in 0.05 M sodium citrate buffer (pH 4.0) was incubated in a water bath for 1 h at 37°C. After the reaction was ended by addition of DNS reagent, the mixture was boiled for 5 min in water for color development and then cooled rapidly. The reducing sugar (liberated) was measured at 540 nm in a UV-VIS spectrophotometer (UV-1601 PC, Shimadzu, Japan). Glucose (Sigma, St. Louis, MO, USA) was used for the standard curve of endoglucanase and one unit of enzyme activity (U/mL) was defined as the amount of enzyme releasing 1 μM of reducing sugar per a minute. All the assays were done in triplicate.
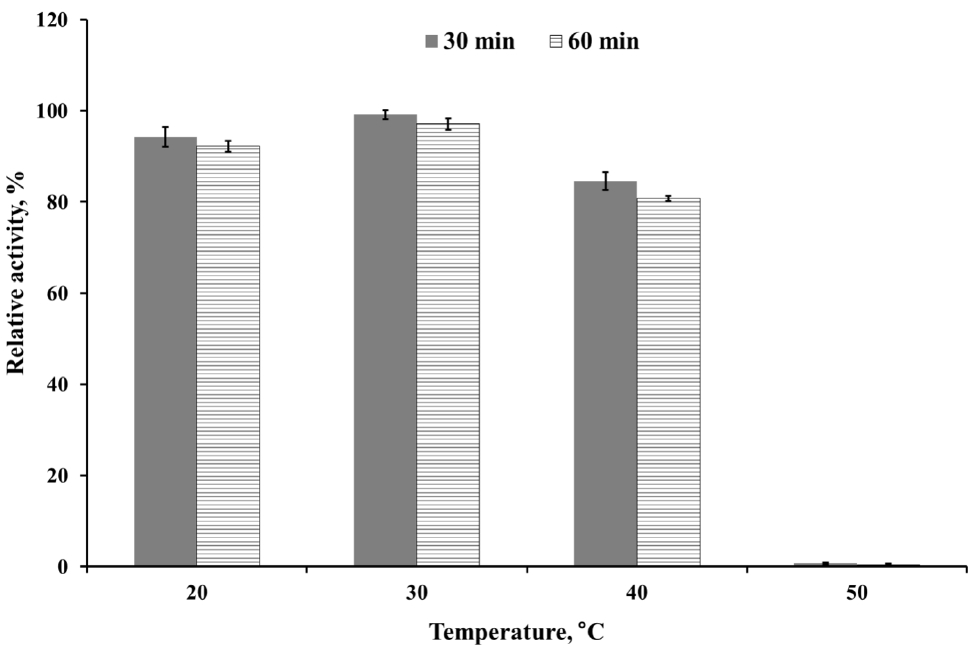
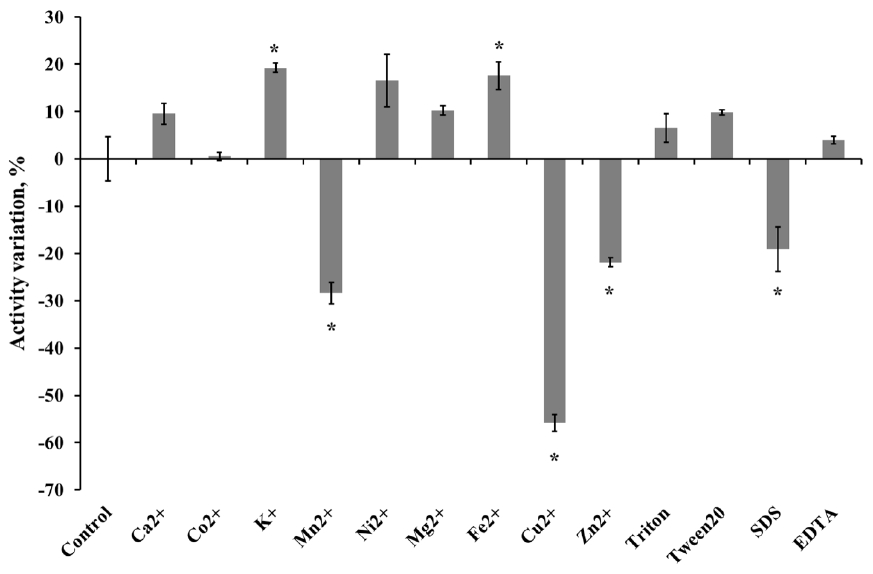
The optimum pH of the purified enzyme was measured in appropriate buffers (0.05 M KCl-HCl, pH 1.0 to 2.0; 0.05 M sodium citrate, pH 3.0 to 5.0; 0.05 M sodium phosphate, pH 6.0 to 8.0; 0.05 M glycine-NaOH buffer, pH 9.0 to 10.0) at 50°C. Enzyme stability at each different pH was determined by pre-incubating the enzyme in the pH buffer solutions as described above at 4°C for 24 h. To determine the optimum temperature for endoglucanase, the purified enzyme preparation was incubated at 20°C to 80°C in 0.05 M sodium citrate buffer (pH 4.0). The effect of temperature on enzyme stability was measured by pre-incubating the enzyme for 30 or 60 min at selected temperatures (20°C to 50°C) then mixed with buffer and CMC solution for 30 min at 50°C. The relative activity of the purified enzyme at various pH or temperature was indicated as the percentage of the maximum enzyme activity at optimal condition. While, the relative activity in stability studies was calculated in contrast to the observed maximum activity at individual pH or temperatures. The influence of chemicals on the enzyme activity was measured after pre-incubating the purified enzyme with 1 mM final concentration of various metal ions (Ca2+, Co2+, K+, Mn2+, Ni2+, Mg2+, Fe2+, Cu2+, and Zn2+) and nonionic reagents (Triton, Tween 20, sodium dodecyl sulfate [SDS], and ethylene diamine tetra acetic acid [EDTA]) in 0.05 M sodium citrate (pH 4) buffer at 4°C for 4 h. Varying the concentration of CMC solution as a substrate from 7.5 to 20 mg/mL was applied to calculate theoretical kinetic values of recombinant Cel5A (rCel5A). The Km and Vmax were calculated using Michaelis-Menten equation at pH 4 and 50°C. All enzyme assays were done in 3 replicates and average values were presented.
The data of enzyme activity were analyzed using the MIXED procedure of SAS (SAS institute Inc., NC, USA) with an appropriate statistical model for each analysis. Differences among different groups were compared with the Tukey’s range test if a significant effect was observed. A statistical significance was declared at p<0.05.
The diluted rumen fluid was spread on the Dehority agar plate contaning 0.5% of Azo-CMC (w/v) and incubated at 39°C for 2 to 3 days. A total of 18 colonies which had clear zones around their area (Figure 1) were randomly selected from the plate and inoculated into the sterilized fresh Dehority liquid medium in Hungate tubes and subcultured. The enzyme activity assay results of final cultivation tubes showed that 8 isolates (1, 2, 3, 6, 12, 16, 17, and 18) had remarkable activity (Figure 2). From those isolates, genomic DNA was extracted and used for the identification study. All isolates had highest similarity (98% to 99%) with E. cellulosolvens Ce2 (Accession number: AY178842.1).
The Cel5A gene′s ORF was amplified from the genomic DNA of isolated E. cellulosolvens using the primers (EC-f, EC-r) which had NdeI, XhoI restriction enzyme sites. The amplified PCR product was digested using adequate restriction enzyme, and ligated with NdeI, XhoI treated pET41b vector (Novagen’s pET Systems, Merck Millipore Corp., Germany) and transformed into E. coli T10F′. The cloned gene was sequenced at NICEM (Seoul National University, Seoul, Korea). The amino-acid sequence was characterized and aligned with the previous published Cel5A gene (Accession number: AB179780.1) using Genbank database (data not shown).
The constructed plasmid was purified and transformed into E. coli BL21 for the production of recombinant enzyme. Isolation of the recombinant clones was done from the LB media containing kanamycin. After colony PCR with the isolated clones, electrophoresis result of the PCR product is presented at Figure 3. The clones having the size 3.5 kb band were selected and used as the host for the expression of the recombinant enzyme. The IPTG induced rCel5A was purified with Ni-NTA His binding resin and used for further characterization.
To determine kinetics of this enzyme, the endoglucanase activity was measured with fixed amount of purified enzyme and 0.5 to 20 mg/mL CMC as a substrate. Theoretical maximal reaction velocities (Vmax) and Michaelis-Menten constant (Km) were calculated against CMC using Lineweaver-Bulk plot (Table 1). Turn over number (Kcat) and catalytic efficiency (Kcat/Km) values of the purified enzyme were 96.69 and 6.88, respectively. The enzyme activity under various conditions was measured to determine the optimal pH, temperature and its stability. The results showed that the purified enzyme had maximum activity at pH 4 and stayed active over pH 3 to 5, showing at least 70% of its optimal activity (Figure 4). It was observed that purified enzyme was highly stable at pH 3 to 9 after 24 h pre-incubation in each pH buffer, and it could maintain its original activity up to almost 90% to 100% (Figure 5). The purified enzyme was active over a broad range of temperature in between 20°C to 80°C. The maximal activity was obtained at temperature of 50°C (Figure 6). However, the enzyme activity dropped sharply as the temperature increased over 40°C after pre-incubation for 30 or 60 min at each temperature (Figure 7). The effect of metal ions and non-ionic reagents after 4 h pre-incubation, is presented in Figure 8. The relative activity was calculated in contrast to the observed maximum activity at no chemical addition. The addition of K+ (potassium), Ni2+ (nickel), and Fe2+ (Iron) cations enhanced enzyme activity by more than 15%. While, the addition of Mn2+ (manganese), Cu2+ (copper), Zn2+ (zinc), divalent cations exhibited negative effect on enzyme activity by about 20% to 50%. Other divalent cations (Ca2+, Co2+, and Mg2+) did not showed any noticeable effect to the enzyme activity. The presence of SDS inhibited the enzyme activity by about 20% while no negative effect was observed with other nonionic reagents.
In this study, we isolated and identified a cultured cellulolytic bacteria from Holstein steer rumens as E. cellulosolvens sp. Among the cellulolytic microorganisms in the rumen, E. cellulosolvens was first isolated by Bryant et al [11] and has been widely observed in Netherlands and South Africa [10]. Many different genes which may have fiber digestion potential, have been cloned from rumen microorganisms. Fibrobacter succinogenes, Butyrivibrio fibrisolvens, Ruminococcus albus, Rumino-coccus flavefaciens, Prevotella ruminicola, and Actinomyces are the small group of microorganisms used for the gene cloning [16]. E. cellulosolvens, one of the major cellulolytic bacteria in the rumen, has not been utilized as a genetic source, although it has many kinds of enzymes and proteins related to the degradation of plant cell wall. In the present study rCel5A from our isolated E. cellulosolvens, encoding endoglucanase, was successfully expressed into E. coli BL21 as the host strain. Yoda et al [17] reported that this gene has tandemly repeated catalytic and cellulose binding domains. Thus, these repeated domains can assist its enzymatic activity and accessibility to the cellulosic materials, and offsets the insufficiency of enzyme productivity of aerobic bacteria by these characteristics.
The purified enzyme has a 14.05 mg/mL Km values. The Km value is a constant number for each given enzyme and enzymes with a small Km value can tightly bind to the substrate. The ability to degrade the defined substrate of purified enzymes can be verified through these kinetic values, including turnover number and catalytic efficiency. Since the catalytic efficiency value is efficient for the direct comparison of effectiveness of each enzyme toward the substrate, this value is used to compare purified enzymes.
The highest activity of the endoglucanase was observed at pH 4, and it can be classified as an acidic endoglucanase. After 24 h pre-incubation in pH 3 to 9 buffer, the enzyme activity remained above 90%, indicating that this enzyme has remarkable stability in acidic conditions. The acidophilic activity of this enzyme might contribute to the cellulose degradation in acidic conditions. In most industrial application cases, it is not economical or efficient to adjust the reaction conditions to neutral pH [18]. Therefore, the rCel5A′s broad range of pH stability might be useful to an industrial application. The purified enzyme had moderate optimal temperature (50°C) but after 30 min pre-incubation over 40°C, enzyme activity was almost removed. If the enzymes were purified from extreme conditions such as hot spring, their optimal activity was observed at 50°C to 60°C and could maintain activity at 65°C, 75°C [19]. Since the recombinant enzyme was originated from the rumen and the rumen has constant temperature range around 40°C, the temperature stability of rCel5A might be reasonable.
Slight increase in enzyme activity occurred by the addition of some cations (K+, Ni2+, and Fe2+). The Mn2+, Cu2+, and Zn2+ inhibited the enzyme activity about 20% to 50%. In many cases, Hg2+ strongly inhibited enzyme activity and Mamo et al [20] reported that Mn2+ inhibited the xylanase activity. But in this experiment, the effect of cations was somewhat different from the above-mentioned experiment. It is not clear, but the cations might have affected the amino acid structure of enzyme active site or other residues [21]. The impact was dependent on several characteristics such as the enzyme’s amino-acid structure, buffer condition and other related characteristics. Triton X-100 and EDTA inhibited endoglucanase (Cel9P) activity of marine bacterium Paenibacillus sp. up to almost 50%. These non-ionic reagents could modify the cellulose surface’s property, so might effect the enzyme activity slightly [15,22].
Eubacterium cellulosolvens was isolated directly from Holstein steer and characterization of the purified enzyme was carried out in detail. The broad pH range, the ability to retain its activity in various chemical sources indicated that this organism and its enzyme may have potential as a candidate for industrial application.
ACKNOWLEDGMENTS
This work was supported by the Rural Development Administration, Republic of Korea (Project No. PJ01285001).
Figure 1
Clear zones around the colonies on solid agar media containing chromogenic carboxymethylcellulose. Cellulolytic microorganism had a colorless zone.
Figure 2
Enzyme activity of isolated microorganisms. The data points and error bars represent the average activities and standard errors of 3 replicates respectively. a–d Values with different scripts did not differ at p<0.05.
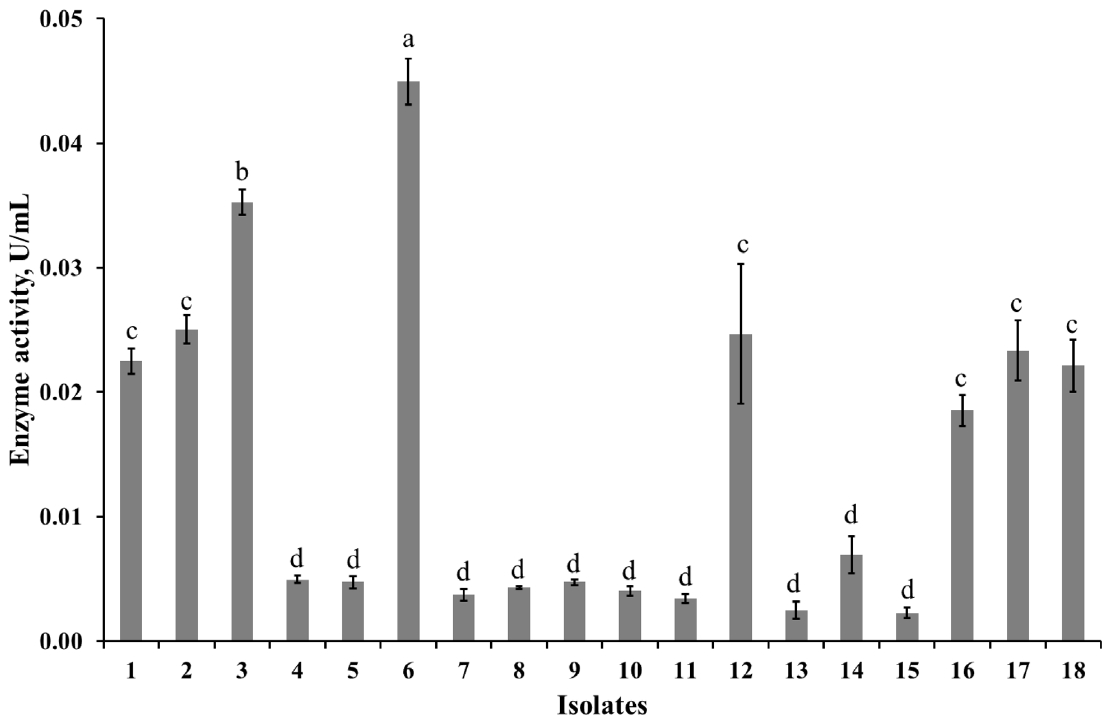
Figure 3
Gel electrophoresis of the colony polymerase chain reaction (PCR) product. M; DNA marker, A to D; colony PCR product of transformants.
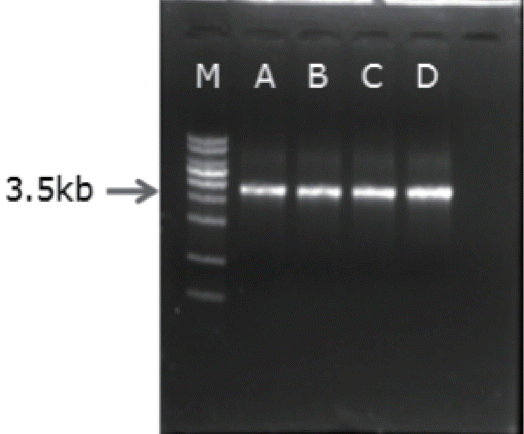
Figure 4
Relative activity of the purified enzyme at various pH at 50°C. The data points and error bars represent the average % and standard errors of 3 replicates respectively. a–i Values with different scripts did not differ at p<0.05.
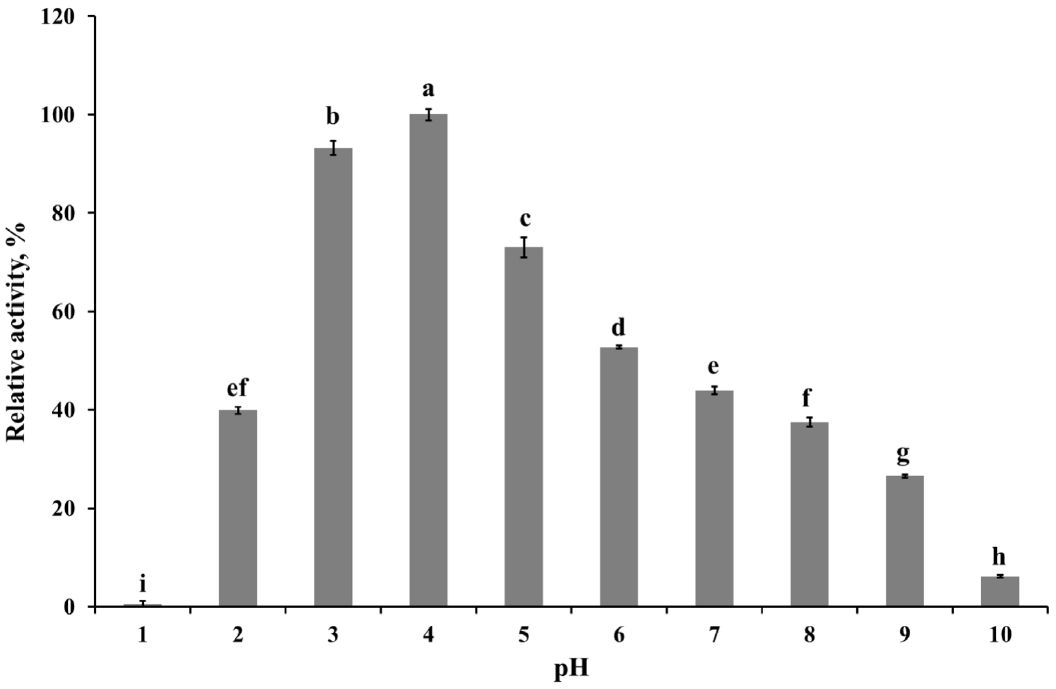
Figure 5
Relative activity of the purified enzyme after 24 h pre-incubation at respective pH. The data points and error bars represent the average % and standard errors of 3 replicates respectively. Values among different pH did not exhibit a significant difference (p = 0.1293).
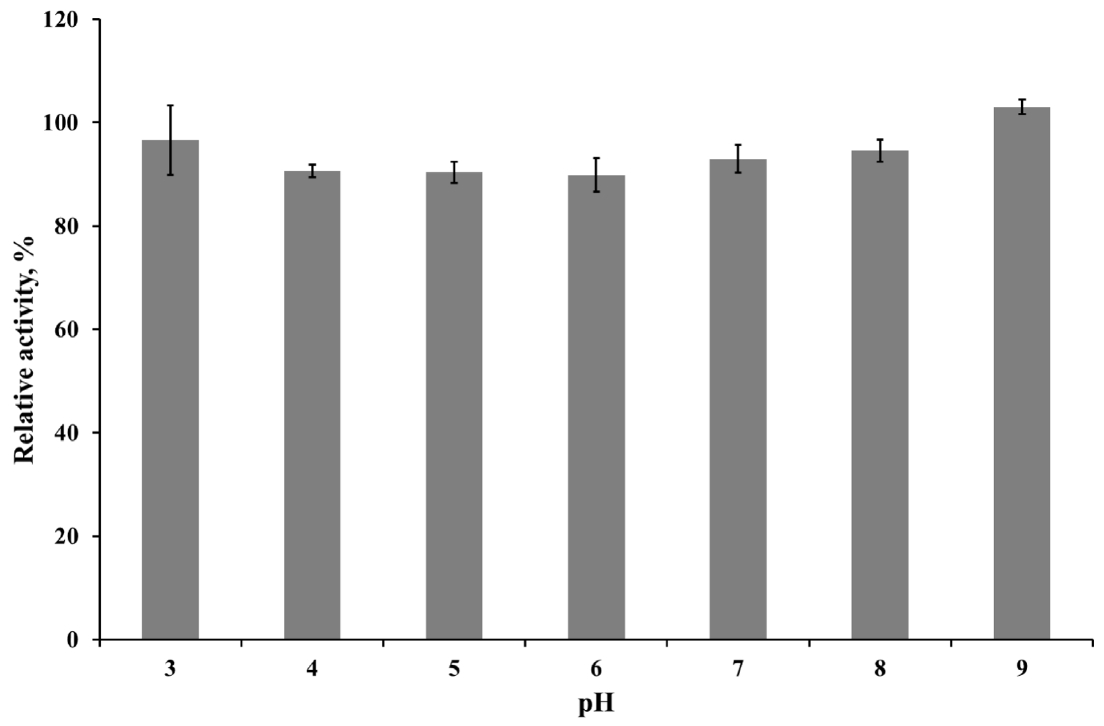
Figure 6
Relative activity of the purified enzyme at various temperature at pH 4. The data points and error bars represent the average % and standard errors of 3 replicates respectively. a–d Values with different scripts did not differ at p<0.05.
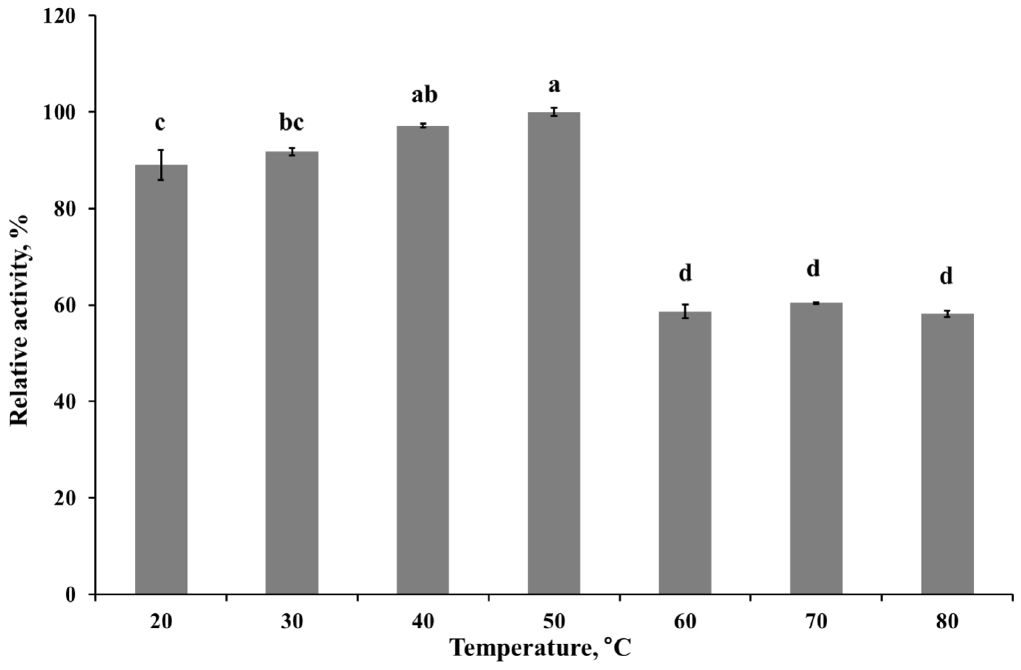
Figure 7
Relative activity of the purified enzyme after 30 or 60 min pre-incubation at respective temperature. The data points and error bars represent the average % and standard errors of 3 replicates respectively. Values between 30 min and 60 min pre-incubation within same temperatures did not exhibit a significant difference (p = 0.6075).
REFERENCES
1. Percival Zhang YH, Himmel ME, Mielenz JR. Outlook for cellulase improvement: Screening and selection strategies. Biotechnol Adv 2006; 24:452–81.


2. Bhat MK, Bhat S. Cellulose degrading enzymes and their potential industrial applications. Biotechnol Adv 1997; 15:583–620.


4. Hungate RE. CHAPTER II-The rumen bacteriaHungate RE, editorThe rumen and its microbes. New York, USA: Academic Press; 1966. p. 8–90.
5. Selinger LB, Forsberg CW, Cheng KJ. The rumen: a unique source of enzymes for enhancing livestock production. Anaerobe 1996; 2:263–84.


6. Krause DO, Denman SE, Mackie RI, et al. Opportunities to improve fiber degradation in the rumen: microbiology, ecology, and genomics. FEMS Microbiol Rev 2003; 27:663–93.



7. Wubah DA, Akin DE, Bomeman WS. Biology, fiber-degradation, and enzymology of anaerobic zoosporic fungi. Crit Rev Microbiol 1993; 19:99–115.


8. Devillard E, Bera-Maillet C, Flint HJ, et al. Characterization of XYN10B, a modular xylanase from the ruminal protozoan Polyplastron multivesiculatum, with a family 22 carbohydrate-binding module that binds to cellulose. Biochem J 2003; 373:495–503.



9. Forsberg C, Cheng KJ. Molecular strategies to optimize forage and cereal digestion by ruminantsBills DD, Kung S-D, editorsBiotechnology and nutrition. Boston, MA, USA: Newnes; 1992. p. 109–47.
10. Prins RA, van Vugt F, Hungate RE, van Vorstenbosch CJ. A comparison of strains of Eubacterium cellulosolvens from the rumen. Antonie Van Leeuwenhoek 1972; 38:153–61.


11. Bryant MP, Small N, Bouma C, Robinson IM. Characteristics of ruminal anaerobic cellulolytic cocci and Cillobacterium cellulosolvens n. sp J Bacteriol 1958; 76:529–37.


12. Scott HW, Dehority BA. Vitamin requirements of several Cellulolytic rumen racteria. J Bacteriol 1965; 89:1169–75.



13. Stackebrandt E, Goodfellow M. Nucleic acid techniques in bacterial systematics. New York, USA: Wiley; 1991.
14. Hofmann MA, Brian DA. Sequencing PCR DNA amplified directly from a bacterial colony. Biotechniques 1991; 11:30–1.

16. Cheng KJ, Lee SS, Bae HD, Ha JK. Industrial applications of rumen microbes - Review. Asian-Australas J Anim Sci 1999; 12:84–92.

17. Yoda K, Toyoda A, Mukoyama Y, Nakamura Y, Minato H. Cloning, sequencing, and expression of a Eubacterium cellulosolvens 5 gene encoding an endoglucanase (Cel5A) with novel carbohydrate-binding modules, and properties of Cel5A. Appl Environ Microbiol 2005; 71:5787–93.



18. Turner P, Mamo G, Karlsson EN. Potential and utilization of thermophiles and thermostable enzymes in biorefining. Microb Cell Fact 2007; 6:9



19. Yang D, Weng H, Wang M, et al. Cloning and expression of a novel thermostable cellulase from newly isolated Bacillus subtilis strain I15. Mol Biol Rep 2010; 37:1923–9.


20. Mamo G, Hatti-Kaul R, Mattiasson B. A thermostable alkaline active endo-β-1–4-xylanase from Bacillus halodurans S7: Purification and characterization. Enzyme Microb Technol 2006; 39:1492–8.







 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print





