Proteomic Assessment of the Relevant Factors Affecting Pork Meat Quality Associated with Longissimus dorsi Muscles in Duroc Pigs
Article information
Abstract
Meat quality is a complex trait influenced by many factors, including genetics, nutrition, feeding environment, animal handling, and their interactions. To elucidate relevant factors affecting pork quality associated with oxidative stress and muscle development, we analyzed protein expression in high quality longissimus dorsi muscles (HQLD) and low quality longissimus dorsi muscles (LQLD) from Duroc pigs by liquid chromatographytandem mass spectrometry (LC-MS/MS)–based proteomic analysis. Between HQLD (n = 20) and LQLD (n = 20) Duroc pigs, 24 differentially expressed proteins were identified by LC-MS/MS. A total of 10 and 14 proteins were highly expressed in HQLD and LQLD, respectively. The 24 proteins have putative functions in the following seven categories: catalytic activity (31%), ATPase activity (19%), oxidoreductase activity (13%), cytoskeletal protein binding (13%), actin binding (12%), calcium ion binding (6%), and structural constituent of muscle (6%). Silver-stained image analysis revealed significant differential expression of lactate dehydrogenase A (LDHA) between HQLD and LQLD Duroc pigs. LDHA was subjected to in vitro study of myogenesis under oxidative stress conditions and LDH activity assay to verification its role in oxidative stress. No significant difference of mRNA expression level of LDHA was found between normal and oxidative stress condition. However, LDH activity was significantly higher under oxidative stress condition than at normal condition using in vitro model of myogenesis. The highly expressed LDHA was positively correlated with LQLD. Moreover, LDHA activity increased by oxidative stress was reduced by antioxidant resveratrol. This paper emphasizes the importance of differential expression patterns of proteins and their interaction for the development of meat quality traits. Our proteome data provides valuable information on important factors which might aid in the regulation of muscle development and the improvement of meat quality in longissimus dorsi muscles of Duroc pigs under oxidative stress conditions.
INTRODUCTION
Variation in meat quality traits is a well-known problem. Meat quality traits are closely related to biological traits of live animal. Hence, biological sciences including genetics, physiology, cell biology, and biochemistry have been widely employed for decades to characterize the biological mechanisms behind major variability of meat quality traits (Bendixen, 2005). Basic knowledge of these mechanisms is essential to reduce the variation in meat quality traits such as tenderness, water-holding capacity, and color. They are also important to understand the physiology of meat animals, especially on muscle growth and development (Lametsch et al., 2002; Hwang et al., 2005). Understanding and changes related to physiochemical factors, genotypes, and many other factors influence postmortem metabolism (Monin et al., 1995; Brocks et al., 1998; Wheeler et al., 2005). Some previous studies have indicated that meat quality is determined by postmortem muscle metabolism (Pette, 2002; Spangenburg and Booth, 2003). At slaughter, muscles become deprived of oxygen as the circulatory system shuts down. This lack of oxygen results in a shift to glycolytic (anaerobic) metabolism and a buildup of lactic acid, causing a drop in muscle pH (Frisby et al., 2005). Accelerated postmortem glycolysis reduces pH and increases temperature within muscle, resulting in excessive protein denaturation and inferior meat quality (Julve et al., 2000). Although extensively researched, the underlying mechanisms of many different meat quality traits are far from well understood due to many factors affecting the quality of meat (Mullen et al., 2006; Hollung et al., 2007). The proteome expressed from the genome is influenced by environmental conditions. Proteome is the molecular link between the genome and the functional quality characteristics of the meat. Therefore, proteomics is a promising and powerful tool in meat science (Lametsch and Bendixen, 2001; Morzel et al., 2004; Jia et al., 2006; Sayd et al., 2006). However proteomics has been, and still are, used in numerous studies on skeletal muscle (Picard et al., 2010).
In this study, we focus on its use in the study of livestock muscle development and meat quality with a focus on the differential expression patterns of proteins and their interactions for the development of meat quality traits.
MATERIALS AND METHODS
Animals and sample collection
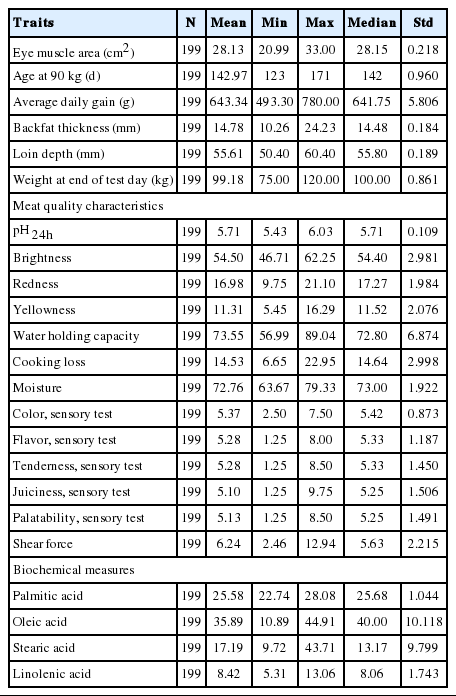
The meat quality characteristics were assessed from 200 randomly selected great grandparent Duroc pigs raised from October 2011 to March 2012 for one production cycle. The live weight ranged from 100 to 120 kg. The carcasses were kept in a freezer (0°C) for 24 h after slaughtering. The frozen carcasses were thawed, deboned, and trimmed. The left side loin was transferred to the laboratory and placed in a deep-freezer (−45°C) for analysis
Gel electrophoresis and silver staining
High quality longissimus dorsi muscles (HQLD) and low quality longissimus dorsi muscles (LQLD) tissues were collected from Duroc pigs. Total protein isolation was performed using PRO-PREP protein extraction solution (iNtRON biotechnology, Sungnam, Korea) according to the manufacturer’s instructions. Concentrations of eluted proteins were measured using Pierce BCA Protein Assay Kit (Thermo scientific, Rockford, IL, USA). Equal amounts of protein samples were precipitated with cold acetone. Protein pellets dissolved in 1× sodium dodecyl sulfate (SDS) sample buffer were separated by 8% and 12% SDS-polyacrylamide gel electrophoresis (PAGE). Following SDS-PAGE, protein spots were visualized using protocols described in PlusOne Silver staining kit (GE Healthcare Bio-Sciences, Uppsala, Sweden). The complete protocol was followed to analyze gels. To prepare gels, the protocol was modified so that glutaraldehyde was omitted from the sensitization step and formaldehyde was omitted from the silver reaction step (Yan et al., 2000). Silver-stained gels were scanned (UMAX PowerLook 2100KL Imaging system, UMAX, Taiwan) and protein profiles were compared.
Liquid chromatography-tandem mass spectrometry
The resulting tryptic peptides were separated and analyzed using reversed-phase capillary high-performance liquid chromatography directly coupled to a Thermo LTQ Orbitrap mass spectrometer using published procedure described by Zuo et al. (2001) with slight modifications. Briefly, a 0.075×20 mm trapping column and a 0.075×120 mm resolving column were packed with C18AQ 218MS low formic acid C18 beads (5 μm in size, 200Å pore size; C18AQ, Michrom BioResources, Auburn, CA, USA) and placed in-line. Peptides were bound to the trapping column for 10 min with 2% (vol/vol) aqueous acetonitrile containing 0.1% (vol/vol) formic acid. The bound peptides were then eluted with a 67 min gradient of 2% to 90% (vol/vol) acetonitrile containing 0.1% (vol/vol) formic acid at a flow rate of 0.2 μL/min. For tandem mass spectrometry, the full mass scan range mode was set at m/z = 50 to 2,000 Da. After determining the charge states of the ion zoom scans, product ion spectra were acquired in MS/MS mode with relative collision energy of 55%. The individual spectra from MS/MS were processed using Protein discoverer 2.1 software (Thermo scientific, USA). The generated peak list files were used to query either the MSDB or the NCBI database using the MASCOT program (http://www.matrixscience.com). We considered modifications of methionine and cysteine, peptide mass tolerance at 2 Da, MS/MS ion mass tolerance at 0.8 Da, allowance of missed cleavage at 2, and charge states (namely, +1, +2, and +3). Only significant hits as defined by MASCOT probability analysis were initially considered.
Cell culture
Mitotic C2C12 mouse myoblasts were obtained from Chonbuk University (Jeonju, Korea). C2C12 were passaged as subconfluent monolayers in growth medium (GM) using Dulbecco’s modified Eagle’s medium (Invitrogen, Grand Island, NY, USA) supplemented with 20% fetal bovine serum, 200 mM L-glutamine, 10 units/mL penicillin, and 10 μg/mL streptomycin. Confluent (90%) myoblasts were differentiated into myotubes by culturing the cells in differentiation medium (DM) with Dulbecco’s modified Eagle’s medium supplemented with 2% horse serum.
3-(4, 5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay
The effects of H2O2 on cell viability were estimated using MTS Assay Kit (Promega, Madison, WI, USA). C2C12 cells were seeded into a 96-well plate for 24 h and treated with H2O2 (12.5 μM to 1 mM) for 24 h or 48 h. MTS solution was added to the plates and incubated at 37°C with 5% CO2 for 2 h. Absorbance at 490 nm was recorded using a GloMax-Multi Microplate Multimode Reader (Promega, USA).
Immunocytochemistry
To detect myosin heavy chain (MYH), myogenic differentiation (MyoD), and myogenin (Myog), cells were blocked with 1% bovine serum albumin and incubated with monoclonal anti-MYH (B-5), anti-MyoD (E-1), or anti-Myog (M-225) antibody at 4°C overnight (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Probed cells were reacted with a 488-conjugated anti-mouse or 594-conjugated anti-rabbit secondary antibody. For nucleus staining, cells were treated with mounting medium with 4′-6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Inc. Burlingame, CA, USA). The cells were visualized using a FluoView confocal laser microscope (Fluoview FV10i, Olympus Corporation, Tokyo, Japan).
Reverse transcription polymerase chain reaction and real-time polymerase chain reaction analysis
Total RNA isolation was performed using TRIzol reagent (Invitrogen, USA) according to the manufacturer's instructions. Briefly, total RNA levels were quantified by absorbance at 260 nm. RNA integrity was evaluated by 1.2% (w/v) agarose gel. Total RNA (2 μg amounts) was reverse-transcribed into cDNA using QuantiTect Reverse Transcription Kit (Qiagen, Chatsworth, CA, USA) according to the manufacturer’s instructions. Real-time polymerase chain reaction analysis (PCR) was performed with SYBR green Premix Ex Taq II (Takara, Dalian, China) using Applied Biosystems StepOne Plus Real-time PCR System (Applied Biosystems, Carlsbad, CA, USA). Relative quantification analysis was performed using the comparative Ct (2 (−ΔΔCT)) method. The expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin was used as endogenous control for the detection of mRNA expression levels. Primers used in the study were listed in Table 4.
Kinetic determination of LDH activity
Commercially available kits for lactate dehydrogenase (DLDH-100, QuantiChrom Lactate Dehydrogenase Kit, Gentaur Molecular, Hayward, CA, USA) were used according to the manufacturers’ instructions (Stentz et al., 2010).
RESULTS AND DISCUSSION
Animals and phenotypes
The average weight before and after slaughtering of the GGP pigs were 98.23 kg and 89.98 kg, respectively. Difference of 10 kg before and after slaughtering was recorded. Important economic traits such as lean percent and eye muscle area showed an average value of 54.74% and 26.03 cm2, respectively (Table 1).
Protein profiles in HQLD and LQLD from Duroc pigs
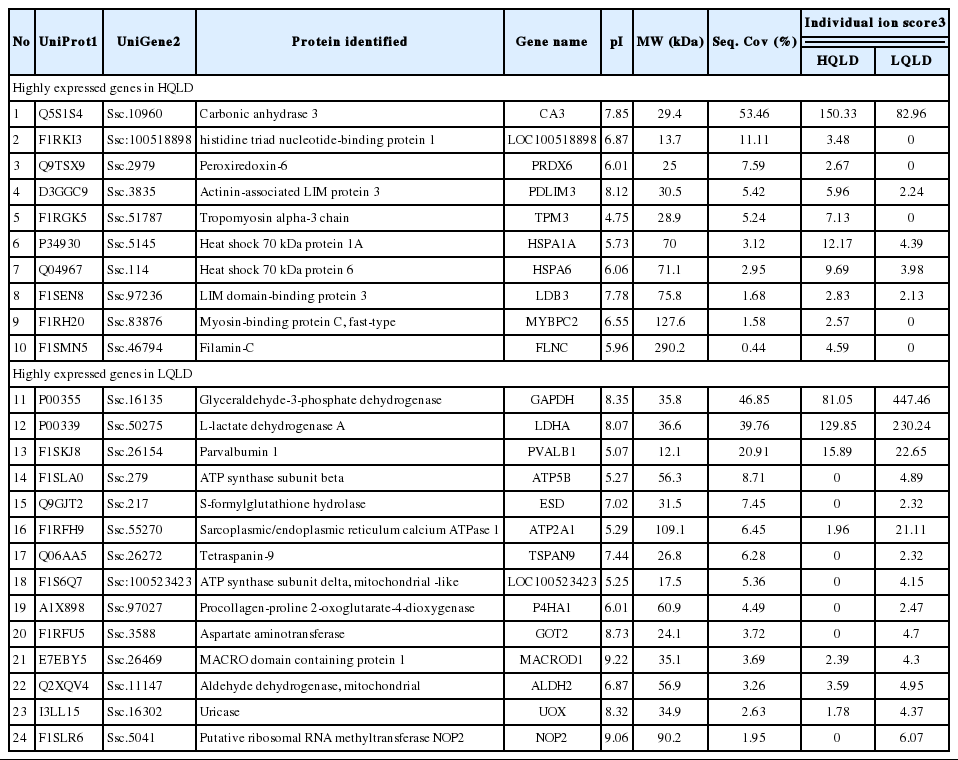
Meat qualities were evaluated by Korea Institute for Animal Products Quality Evaluation (KAPE) authorized by South Korean government to perform animal products grading service. The normality test was applied to show normal distribution of the traits. The highest and lowest meat grades for pH and water holding capacity were identified from the sample which accounted for 10% (20 heads) of the total population (Table 2). To obtain a comprehensive overview of protein components in HQLD and LQLD from 12 individuals, protein profiles of whole lysate of HQLD and LQLD separated by 8% and 12% SDS-PAGE were assessed by silver-stained image analysis (Figure 1A). Patterns of total protein components in whole lysate of HQLD and LQLD were similar among the individual six groups. However, significantly fewer proteins (i.e. band a and b) were expressed in HQLD compared to in LQLD. Two different protein spots were identified by a mass spectrometric analysis. Myosin binding protein C (MYBPC2) expressed during skeletal muscle development (Gurnett et al., 2010) had higher expression in HQLD than in LQLD. However, lactate dehydrogenase A (LDHA) catalyzing the conversion of pyruvate to lactate during glycolysis (Fan et al., 2011) had higher expression in LQLD than in HQLD (Figure 1B).
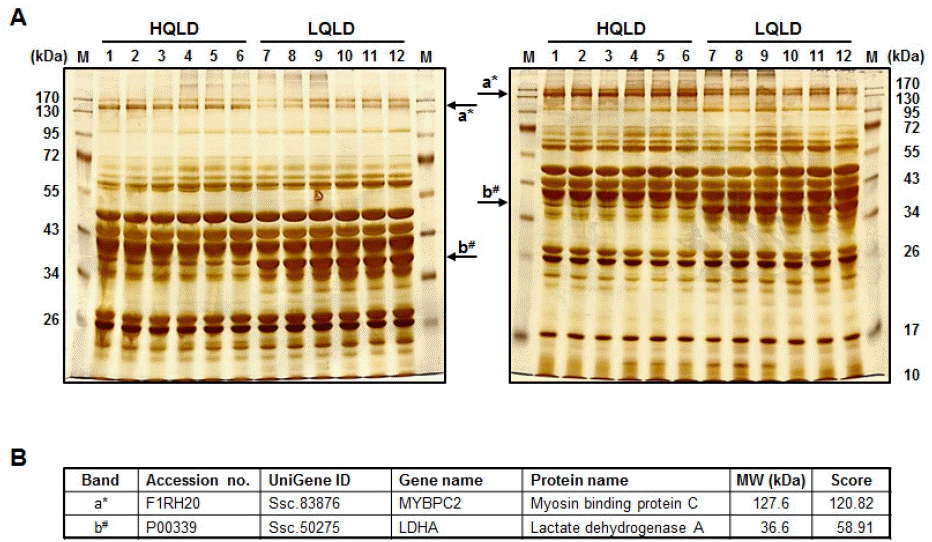
Protein profiles of high quality longissimus dorsi muscles (HQLD) and low quality longissimus dorsi muscles (LQLD) from Duroc pigs by image analysis. (A) The overall patterns of total protein bands from individuals. All gels were visualized by sliver staining. (B) Two different protein spots were identified by a mass spectrometric analysis. a*: MYBPC2, myosin binding protein C; b#: LDHA, lactate dehydrogenase A.
Protein identification and gene ontological classification by LC-MS/MS-based proteomic analysis
Next, we performed liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based proteomic analysis to elucidate the proteins involved in longissimus dorsi muscle (i.e. HQLD and LQLD) properties involved in meat quality. Among the total 24 proteins identified, 10 and 14 proteins were confirmed to be highly expressed in HQLD and LQLD, respectively. All identified proteins were clustered into the following seven categories (Figure 2A) based on information obtained from DAVID gene ontology (GO) database (http://david.abcc.ncifcrf.gov) and UniProt (http://www.uniprot.org): Catalytic activity (31%), ATPase activity (19%), oxidoreductase activity (13%), cytoskeletal protein binding (13%), actin binding (12%), calcium ion binding (6%) and structural constituent of muscle (6%) (Supplementary Table S1). The GO analysis was performed using DAVID Bioinformatics Resources 6.7 categories for both molecular function (MF) and biological process (BP). Depending on the MF in which the proteins were involved, they were categorized into the following three groups (Figure 2B): Cytoskeletal protein binding (40%), actin binding (40%), and structural constituent of muscle (20%) (Supplementary Table S2). Depending on the BP in which the proteins were involved, they were categorized into the following five groups (Figure 2C): Primary metabolic process (26%), cellular metabolic process (26%), catabolic process (18%), nitrogen compound metabolic process (19%), and oxidation reduction (11%) (Supplementary Table S3). The expression changes of the up- and down-regulated proteins in HQLD and LQLD of Duroc pigs were summarized in Table 3. LDHA was selected and subjected to further analysis by LDH activity assay and in vitro study of myogenesis under oxidative stress conditions.
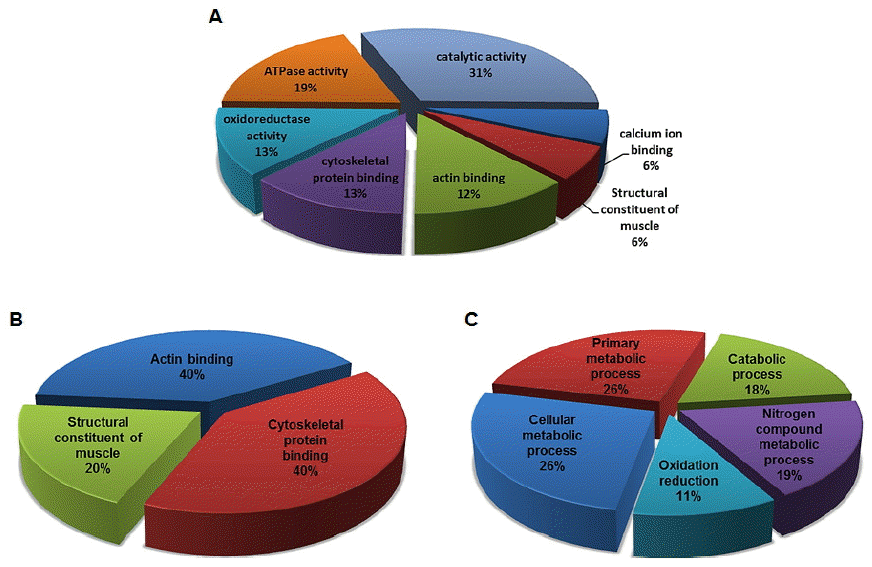
Ontological classifications of differentially regulated proteins in high quality longissimus dorsi muscles (HQLD) and low quality longissimus dorsi muscles (LQLD) from Duroc pigs. Of the total 24 identified proteins, 10 and 14 proteins were highly expressed in HQLD and LQLD, respectively. (A) The identified proteins were clustered into 7 categories based on information obtained from DAVID gene ontology (GO) database; (B) The identified proteins were clustered into 3 categories based on their molecular function; (C) The identified proteins were clustered into 5 categories based on their biological processes.
Gross changes in C2C12 myoblasts in response to myogenic differentiation
C2C12 myoblasts serve as an experimentally tractable model system for investigating the molecular basis of skeletal muscle cell specification and development (Kislinger et al., 2005). A temporally well-defined myogenic differentiation program can be selectively triggered in cultured C2C12 myoblasts upon withdrawal of GM and mitogens (Gramolini and Jasmin, 1999). When switched to DM, mitotic C2C12 myoblasts rapidly cease proliferation and initiate a synchronously terminal differentiation program (Figure 3A). To investigate the patterns of protein expression and efficiency of myotube formation during myogenesis under oxidative stress condition, undifferentiation C2C12 cells were treated with various concentrations of H2O2 (12.5 μM to 1 mM). Cytotoxicity was negligible with 200 μM H2O2. However, up to 1 mM H2O2 did reduce viability which was confirmed by 3-(4, 5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) (MTS) assay (Figure 3B). Control cells exhibited striking morphological changes over the course of 3 to 7 days, eventually fusing into mature multinucleated myotubes (i.e. by day 5). However, H2O2-treated cells exhibited thread-like shape without fusing into mature multinucleated myotubes (Figure 3C). Myotubes were identified by immunocytochemistry with anti-myosin heavy chain (MYH) antibody (Figure 3D).
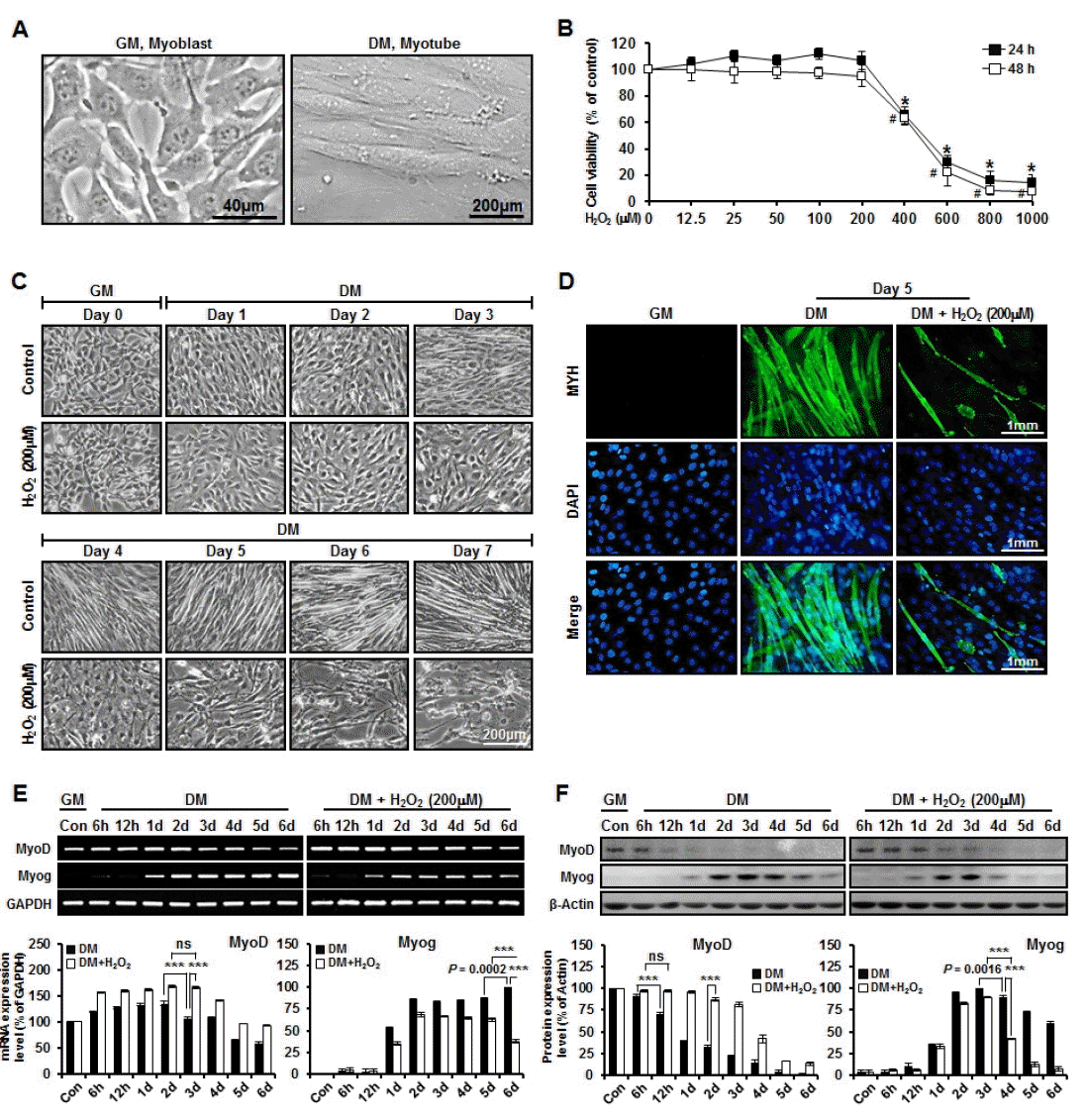
Gross changes and cell viability in myogenic differentiation under oxidative stress condition. (A) Morphological changes of myoblast during myogenesis. Arrow head and dashed lines indicating myoblast with mono nucleus and myotube with three or more nuclei, respectively; (B) Cytotoxic effects of H2O2 in C2C12 treated with the various concentrations of H2O2 (12.5 μM to 1 mM). Data represented mean value of three independent experiments; bars, standard deviation. * Significantly different compared to untreated conditions (24 h); #, significantly different compared to untreated conditions (48 h) (n = 3, p<0.0001); (C) Light microscopy-based images of undifferentiated (day 0) proliferating C2C12 myoblasts and differentiating cells at various time points (up to day 7) under normal and oxidative stress conditions (H2O2 [200 μM]); (D) Immunocytochemistry presented the MYH expression levels of undifferentiated (day 0) and differentiated (day 5). GM, growth medium; DM, differentiation medium. (E) Oxidative stress induced down-regulation of Myog gene expression and continuous up-regulation of MyoD gene expression with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a control; (F) Oxidative stress induced down-regulation of Myog protein expression and continuous up-regulation of MyoD protein expression with β-actin as a control. Student’s t test was performed to evaluate statistical significance (ns, non-significant; *** p<0.0001; mean±standard error of the mean; n = 3).
In vitro model of myogenesis under oxidative stress condition
The expression of myogenic regulatory factors consisting of MyoD and Myog characterizes various phases of skeletal muscle development, including myoblast proliferation, cell-cycle exit, cell fusion, and the maturation of myotubes to form myofibers (Lee et al., 2014). MyoD, the chief regulatory molecule of myogenic differentiation (Langen et al., 2004), plays an important role in cell cycle exit of differentiating myoblasts (Guo et al., 1995; Halevy et al., 1995) Terminal differentiation of myoblast, driven by expression of Myog, is essential for the formation of functional multinucleated myofibers (Liu et al., 2012). In order to study the patterns of MyoD and Myog expression (gene and protein levels) depending on oxidative stress condition during myogenesis, we used western blot and RT-PCR analyses. Our data revealed that, among various genes subjected to comparison, MyoD had significant (p<0.0001) higher expression under oxidative stress condition than under normal condition on day 3. The expression of MyoD on day 3 was not significantly different from that on day 2 under oxidative stress condition. Myog had significant (p<0.0001) higher expression under normal condition than under oxidative stress condition at day 6. Under oxidative stress condition, the expression of Myog was significantly (p<0.05) decreased at day 6 compared to that at day 5 (Figure 3E). The mRNA expression levels for selected genes were analyzed by quantitative real-time PCR with specific primer (Table 4). Western blot analysis data revealed that, of different proteins subjected to comparison, MyoD had significantly (p<0.0001) higher expression under oxidative stress condition than under normal condition at day 2. Under oxidative stress condition, the expression of MyoD at 12 h was not significantly different from that at 6 h. Moreover, Myog had significantly (p<0.0001) higher expression under normal condition than under oxidative stress condition at day 4. Under oxidative stress condition, the expression of Myog was significantly more decreased (p = 0.0016) at day 4 than at day 3 (Figure 3F). These results indicate that H2O2-induced oxidative stress inhibits myogenesis through the down-regulation of Myog and the continuous up-regulation of MyoD.
Relationships between LDHA gene expression and myogenesis
Previous studies reported that porcine myogenic differentiation 1 (MyoD1) gene has been mapped to swine chromosome 2p14-p17 which is involved in the regulation of the proliferation and differentiation of skeletal muscle cells. LDHA genes mapped close to MyoD are involved in energy metabolism and protein transport processes. LDHA genes might play important roles in muscle development (Qiu et al., 2010). However, little is known about porcine LDHA genes. Therefore, we determined the relationships between LDHA gene expression and myogenesis under normal and oxidative stress condition. To investigate whether oxidative stress regulated LDHA expression at genetic levels, we used quantitative real-time PCR. The mRNA expression levels of selected genes were subjected to quantitative real-time PCR with specific primers (Table 4). Figure 4A showed that LDHA genes were increased up to day 5 during myogenesis under normal and oxidative stress condition. LDHA expression was significantly (p = 0.0088) higher under oxidative stress condition. These results indicate that up-regulated LDHA genes induced by oxidative stress might play dysfunctional roles in myogenesis.
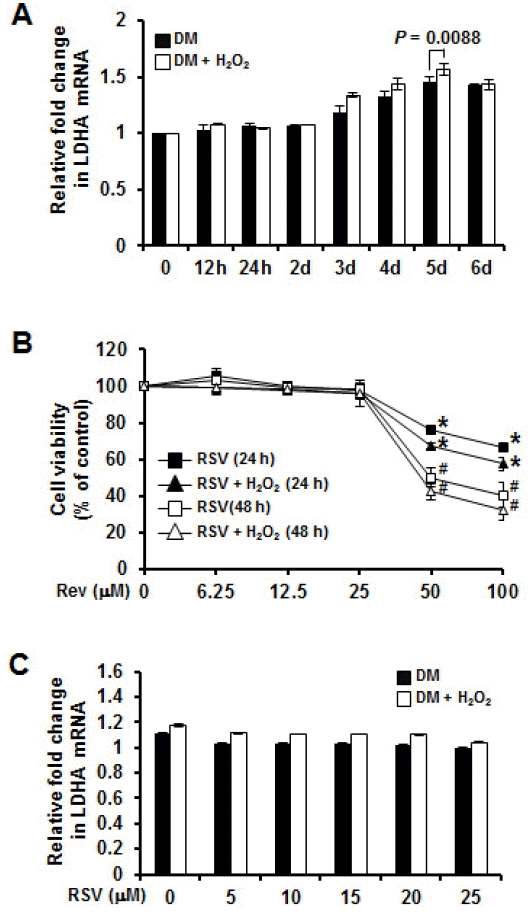
Gene and protein expression levels on high quality longissimus dorsi muscles (HQLD) and low quality longissimus dorsi muscles (LQLD) depending on oxidative stress. Myoblast and myotube cDNA and protein were determined by real-time PCR analysis and western blot analysis, respectively. (A) The quantitative differences of lactate dehydrogenase A (LDHA) at the transcriptional level were measured by real-time PCR during myogenesis with β-actin as control; (B) Cytotoxic effects of resveratrol (RSV) in C2C12 treated with various concentrations of RSV (6.25 μM to 100 μM). Data represented mean value of three independent experiments; bars, standard deviation. * Significantly different compared with untreated conditions (24 h); #, significantly different compared to untreated conditions (48 h) (n = 3, p<0.0001); (C) Quantitative differences of LDHA at the transcriptional level were measured by real-time PCR in RSV-treated C2C12 during myogenesis.
Antioxidant properties of resveratrol
Resveratrol (RSV), a well-known phytocompound and food component, has antioxidative and multifunctional bioactivities (Wu et al., 2013). Previous studies have reported that RSV in skeletal muscle acts on protein catabolism and muscle function and confers resistance against oxidative stress, injury, and cell death. However, its action mechanisms and protein targets in myogenesis process are not completely understood (Montesano et al., 2013). Therefore, we determined the effect of RSV on LDHA gene expression in myogenesis under oxidative stress condition. C2C12 cells were treated with various concentrations of RSV (6.25 μM to 100 μM). Cytotoxicity was negligible with RSV (25 μM) under normal and oxidative stress conditions. However, up to 100 μM RSV did reduce viability which was confirmed by MTS assay (Figure 4B). However, after RSV treatment, there was no significant difference in mRNA expression of LDHA between normal condition and oxidative stress condition (Figure 4C).
Confirmation of lactate dehydrogenase activity
Elevation of plasmatic LDH levels are characteristic responses to strenuous exercise which are often used as indicators of muscle damage (Kanter et al., 1988; Bouzid et al., 2014). However, difference of LDH activity between HQLD and LQLD of Duroc pigs is not well determined. Our data showed that LDH activity was significantly (p = 0.0003) higher in LQLD than in HQLD of Duroc pigs (Figure 5A). Moreover, higher LDH activity was positively correlated with in vitro model of myogenesis under oxidative stress condition. In addition, LDH activity was significantly reduced by RSV treatment (Figure 5B). We also confirmed the patterns of MyoD and Myog expression under oxidative stress condition and RSV treatment during myogenesis by immunocytochemistry. Oxidative stress induced down-regulation of Myog and continuous up-regulation of MyoD. The down-regulation induced by oxidative stress was recovered by RSV treatment. The up-regulation of MyoD induced by the oxidative stress was reduced by the treatment of RSV in myogenesis (Figure 5C). In addition, there was a significant correlation between MyoD and Myog expression (gene and protein levels) under oxidative stress condition during myogenesis (Figure 5D). These results indicate that high activity of LDH by oxidative stress will result in dysfunction of myogenesis and that RSV treatment will result in its functional recovery.
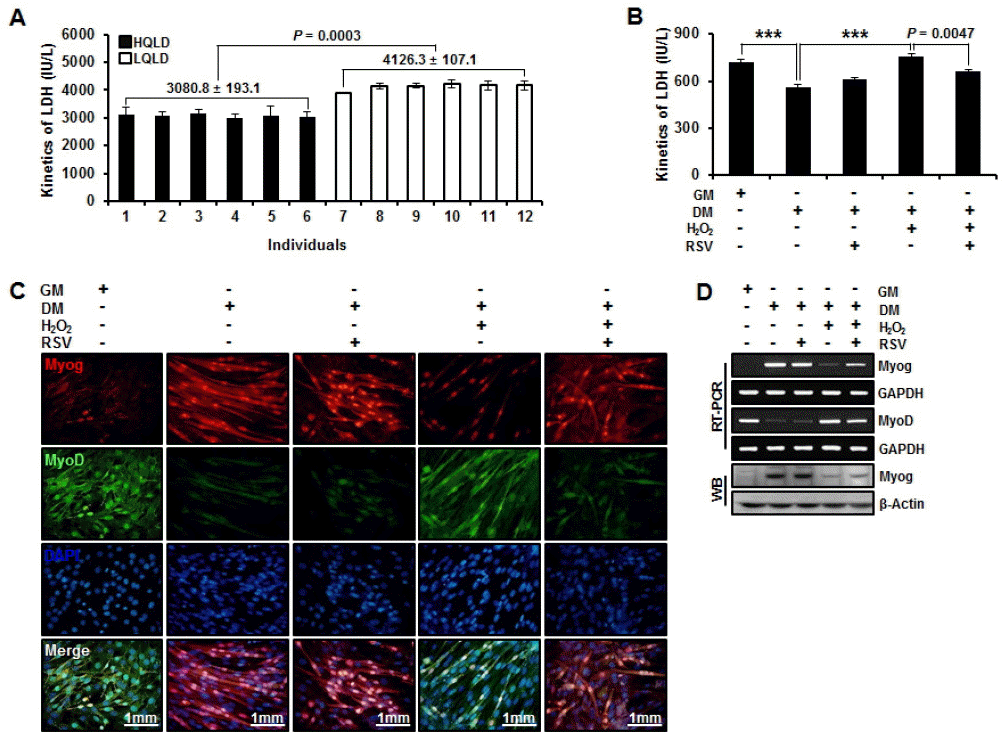
Antioxidant effect of resveratrol in lactate dehydrogenase activity and myogenesis. (A) Difference of lactate dehydrogenase (LDH) activity between high quality longissimus dorsi muscles (HQLD) and low quality longissimus dorsi muscles (LQLD) from individual six groups of Duroc pigs. (B) In vitro model of myogenesis under oxidative stress condition and changes of lactate dehydrogenase activity by resveratrol (RSV) treatment. Student’s t test was performed to evaluate statistical significance (*** p<0.0001; mean±standard error of the mean; n = 3); (C) Patterns of myogenic differentiation (MyoD) and myogenin (Myog) expression depending on oxidative stress condition and RSV treatment during myogenesis by immunocytochemistry; (D) Patterns of MyoD and Myog expression (gene and protein levels) depending on oxidative stress condition and RSV treatment during myogenesis by reverse transcription polymerase chain reaction and western blot analysis.
In conclusion, our data demonstrated that up- or down-regulation of genes and proteins are involved in muscle development, muscle function, actin organization, oxidative stress, cell proliferation, cell differentiation, and cell growth. In this paper, differential expression patterns of genes and their interaction are found to be important for the development of meat quality traits. Our proteome data provided valuable information on differentially expressed genes (LDHA) and activity of LDH in HQLD and LQLD from Duroc pigs, which may aid in the regulation of muscle development. Our study provided experimental evidence for RSV as an important regulator to improve meat quality grades in porcine. However, further study is required to determine the relationship between differential expression of genes or proteins and their direct effects on meat quality.
Supplemental Table
ACKNOWLEDGMENTS
This work was carried out with the support of “Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ011682)” Rural Development Administration, Republic of Korea.
Notes
CONFLICT OF INTEREST
We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.