State of the art on the physical mapping of the Y-chromosome in the Bovidae and comparison with other species — A review
Article information
Abstract
The next generation sequencing has significantly contributed to clarify the genome structure of many species of zootechnical interest. However, to date, some portions of the genome, especially those linked to a heterogametic nature such as the Y chromosome, are difficult to assemble and many gaps are still present. It is well known that the fluorescence in situ hybridization (FISH) is an excellent tool for identifying genes unequivocably mapped on chromosomes. Therefore, FISH can contribute to the localization of unplaced genome sequences, as well as to correct assembly errors generated by comparative bioinformatics. To this end, it is necessary to have starting points; therefore, in this study, we reviewed the physically mapped genes on the Y chromosome of cattle, buffalo, sheep, goats, pigs, horses and alpacas. A total of 208 loci were currently mapped by FISH. 89 were located in the male-specific region of the Y chromosome (MSY) and 119 were identified in the pseudoautosomal region (PAR). The loci reported in MSY and PAR were respectively: 18 and 25 in Bos taurus, 5 and 7 in Bubalus bubalis, 5 and 24 in Ovis aries, 5 and 19 in Capra hircus, 10 and 16 in Sus scrofa, 46 and 18 in Equus caballus. While in Vicugna pacos only 10 loci are reported in the PAR region. The correct knowledge and assembly of all genome sequences, including those of genes mapped on the Y chromosome, will help to elucidate their biological processes, as well as to discover and exploit potentially epistasis effects useful for selection breeding programs.
INTRODUCTION
In the last years, the genome of different farm animals, including pig [1], cow [2], buffalo [3], sheep [4], goat [5], horse [6], and camel [7], has been sequenced. Furthermore, there are species like the alpaca that have progressed slowly in the assembly because of their difficult karyotype [8,9]. The data obtained from these studies have significantly contributed to the understanding of domestication [10], to the selection of better breeds [11,12] and to the interaction between genetic traits and the environment [13,14].
In the studies of mammalian genome and in particular of livestock species, the sex chromosomes (X and Y) have considerable importance. Especially the Y-chromosomes (Chr Y), which have undergone substantial evolutionary changes and losing about 95% of the ancestral genesis [15]. The Chr Y is the smallest chromosome and consists of 2% to 3% of the haploid genome and, according to the species, may contain between 70 and 200 genes. It is involved in the segregation of the sex chromosomes in male meiosis. Therefore, it plays a key role for evolutionary studies, speciation, male infertility and/or subfertility due to its unique features, such as long non-recombining regions (NRYs), abundance of repetitive sequences, and holandric inheritance pattern [16]. This chromosome is generally separated into two distinct domains: the pseudoautosomal region (PAR) and the non-pseudoautosomal region, also known as the male-specific region of the Y chromosome (MSY). The PAR is a characteristic DNA region distinct from the autosomes that exhibits sequence homology between sex chromosomes [17].
The MSY region is not subject to pairing during meiosis. Therefore, it has been considered a NRY, although abundant recombination has been reported in humans [18,19]. Furthermore, the MSY contains gene families encoding multi-copy proteins associated with male fertility [15,20].
To date, Chr Y sequencing has been completed and characterized only in some species, including the human [18], the chimpanzee, the rhesus macaque [21], the mouse [22], the pig [23] and the horse [24]. Conversely, data are not available for other species.The molecular cytogenetic techniques, like the fluorescence in situ hybridization (FISH), offer a powerful tool in support of the bioinformatics pipelines following next generation sequencing projects. In fact, genomes assemblies are prone to errors, as they have been detected for instance in cattle, goats and sheep genomes by FISH [25,26]. Therefore, the use of the cytogenetic approaches are still fundamental for a correct assembly process. In fact, the FISH by bacterial artificial chromosome (BAC) clones allows assigning sequence contings to specific chromosomes and allows establishing the correct physical order of each DNA fragment, as demonstrated for both animal and vegetable sequencing processes [27,28].
Since the knowledge of the genomes elevates to a different meaning if associated with chromosomes, both for the accuracy of the information and for the evaluation of all related biological aspects [29]; in the present study, we describe the current state of physical gene mapping by FISH in the main species of zootechnical interest (cattle, buffalo, sheep, goat, pig, horse, and alpaca). The choice to focus the attention on this point lies in the current gap of genome assembly data on Chr Y and in the fundamental importance of cytogenetics in the study and interpretation of the genome [30,27,31], as already understood in 1920 by Hans Winkler when he coined the term “genome”.
BOVIDS Y-CHROMOSOME AND PSEUDOAUTOSOMAL REGION
The Bovidae, in particular we considered Bos taurus (BTA), Bubalus bubalis (BBU), Ovis aries (OAR) and Capra hircus (CHI), play a role of fundamental importance for the livestock sector from an economic point of view. For this reason, these species have been investigated deeply from a genetic point of view, in particular for their fertility and consequently their sex chromosomes [32–35].
Although generally small, the Y-chromosome has different size and shape in the bovids. Indeed, in BTA the Chr Y is a small submetacentric, in BBU is a small acrocentric, while in both OAR and CHI is a very small metacentric [36,37]. However, in some breeds of bovids, the Chr Y is fused with an autosome, as in the case of the male of Gazella granti [38]. Moreover, the Chr Y shows few bands because it is almost completely heterochromatic. Besides, the pseudoautosomal boundary (PAB) separates the PAR with similar sequences between X and Y-chromosomes, from the MSY of reduced homology and specific to individual sex chromosomes [39]. In bovids, the genes present in the PAR of the Chr Y are the same in content and sequence as in the X chromosome [15].
Bos taurus
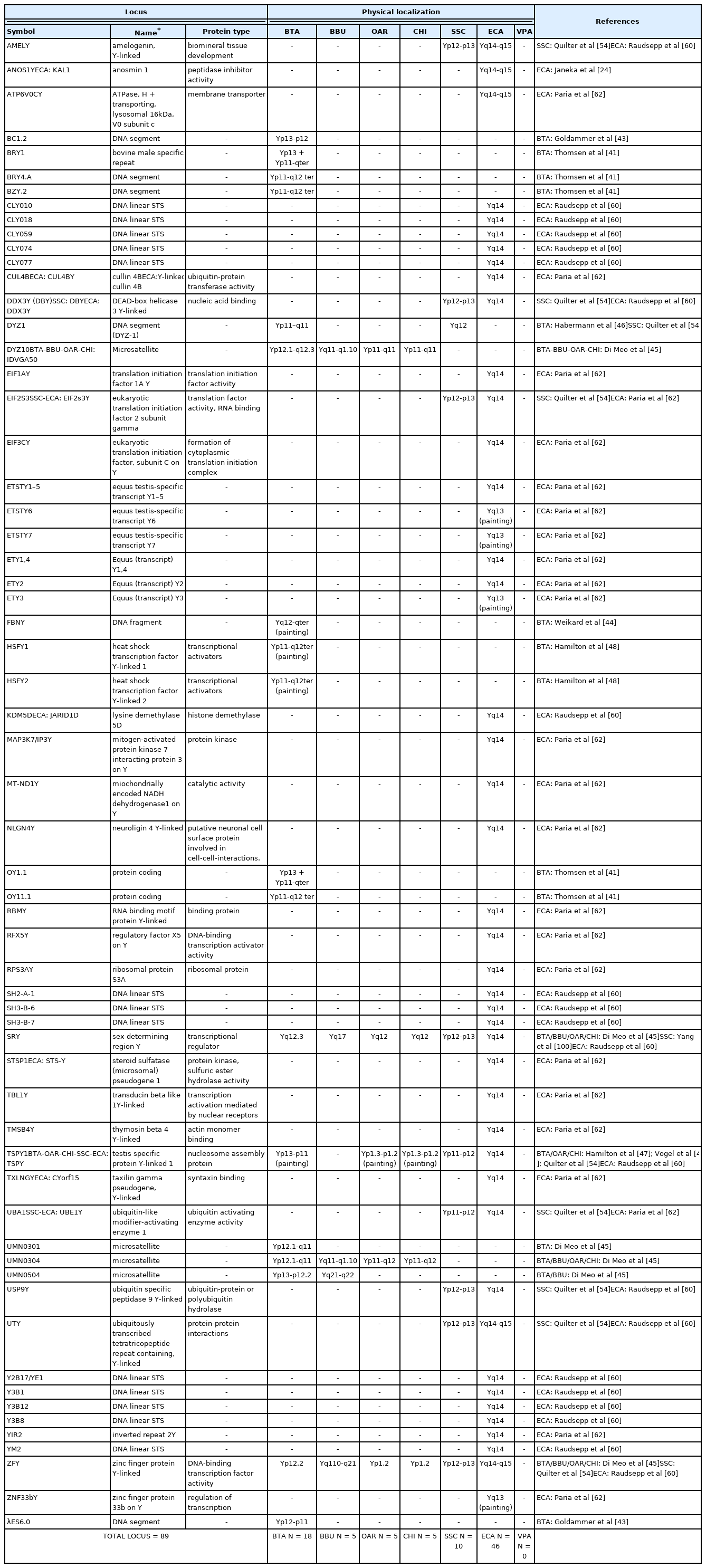
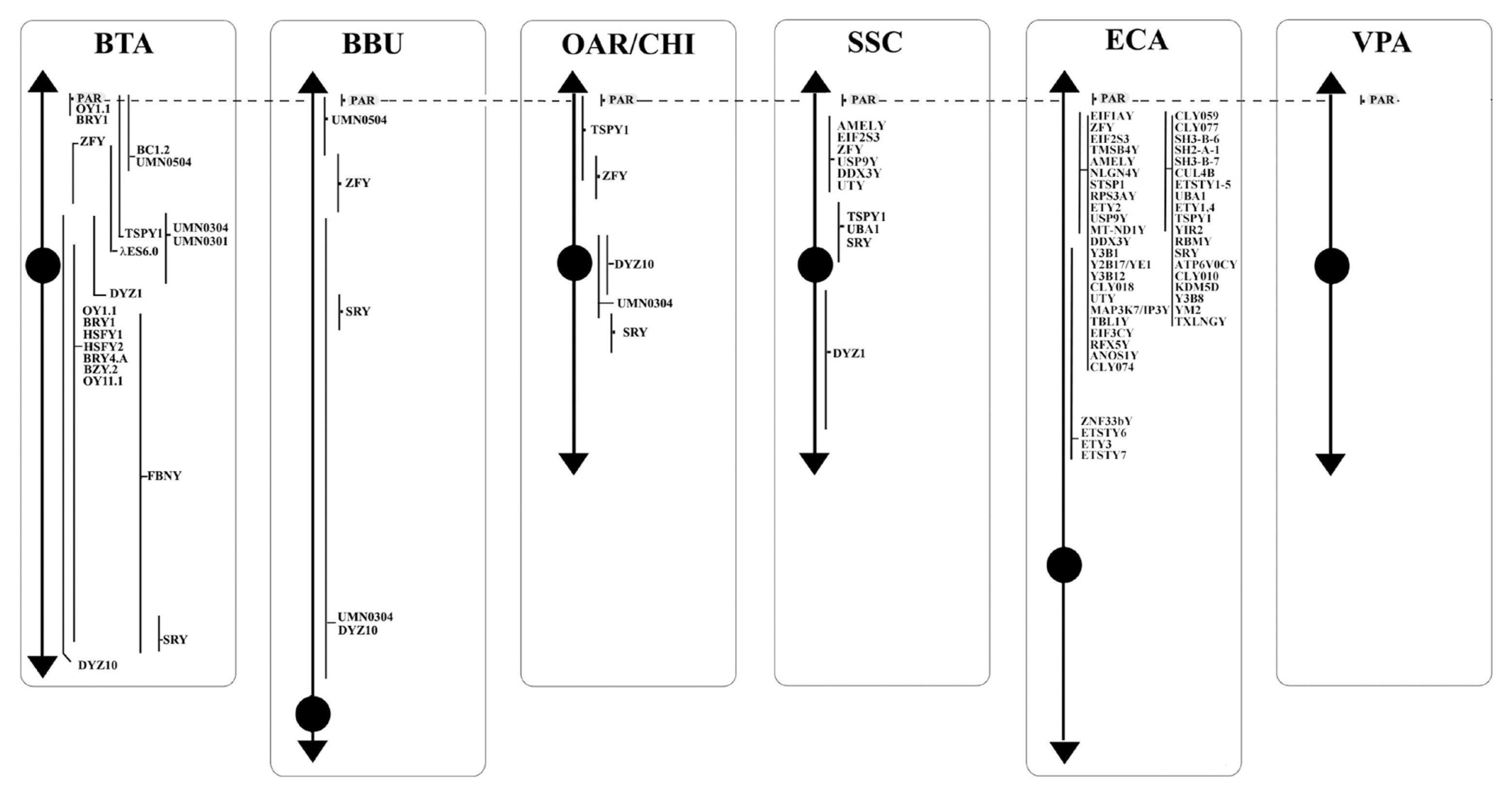
The bovine genome was among the first genomes to be partially sequenced, after the sequencing of the human one. The latest cattle genome assembly release (ARS-UCD1.2) of the whole genome, contains 29 pairs of the autosomes, the X chromosome, and unplaced sequences. Information on the Chr Y is not available yet. Through the years, only few studies focused on the physical mapping of molecular markers by FISH [40–48]. These studies allowed confirming the presence and the precise location of eighteen genes on the MSY, as showed in Table 1 and Figure 1.
Cytogenetic chromosome localisation of physically mapped MSY loci on the Y-chromosome of BTA, BBU, OAR, CHI, SSC, ECA, and VPA and their references
Schematic representation of MSY loci along the Y chromosomes of BTA, BBU, OAR, CHI, SSC, ECA, and VPA. MSY, male-specific region of the Y chromosome; BTA, Bos taurus; BBU, Bubalus bubalis; OAR, Ovis aries; CHI, Capra hircus; SSC, Sus scrofa; ECA, Equus caballus; VPA, Vicugna pacos.
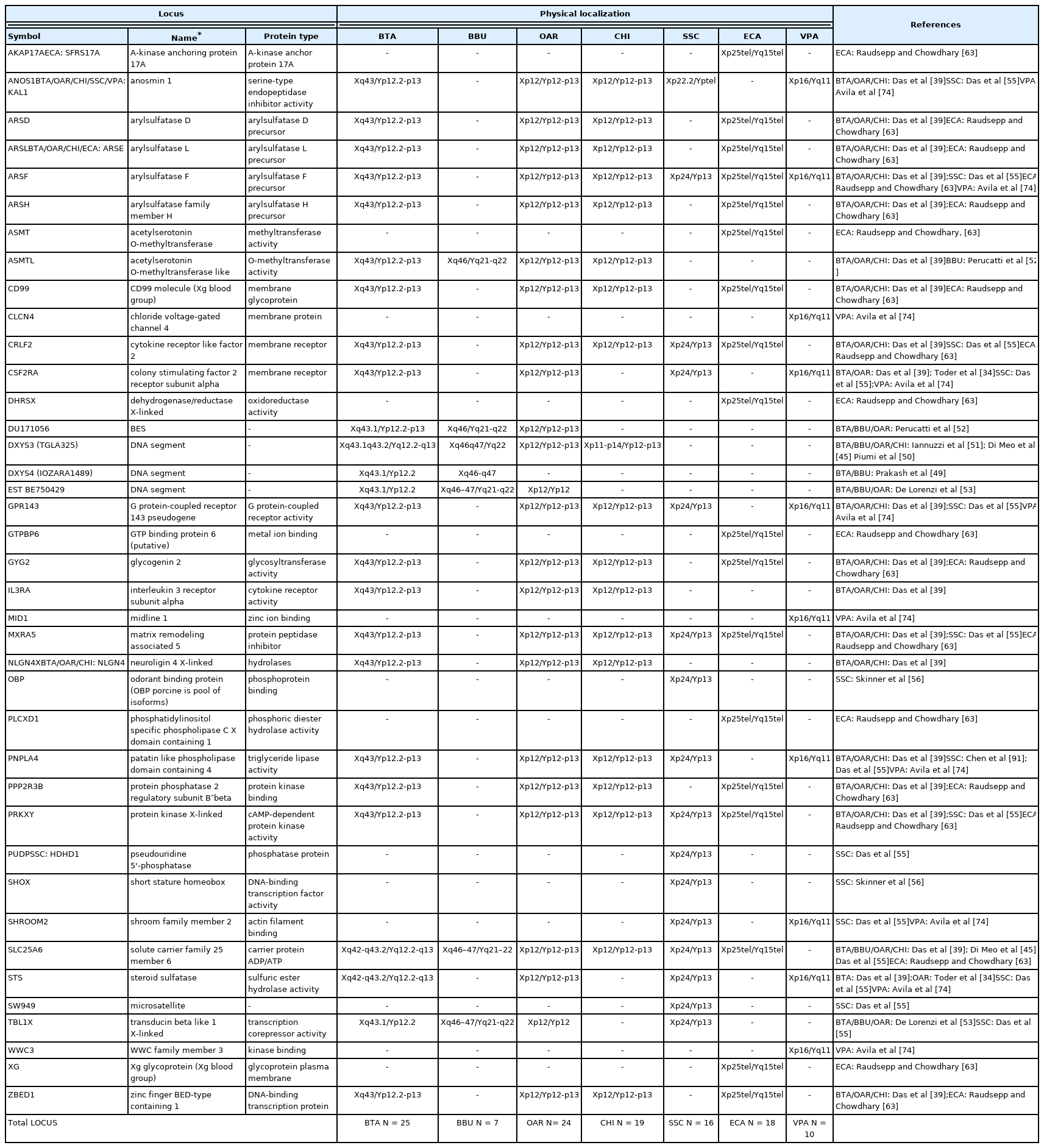
The PAR region is located at the telomere of the short arm of the Chr Y and contains twenty five genes physically mapped [34,49–51,45,39,52,53], as showed in Table 2 and Figure 2.
Cytogenetic chromosome localisation of physically mapped PAR loci on the Y-chromosome of BTA, BBU, OAR, CHI, SSC, ECA and VPA, and their references
Bubalus bubalis
The buffalo genome has been recently sequenced with a contiguity surpassing both human and goat genomes [3]. The assembly UOA_WB_1 containis 24 pairs of the autosomes, the X chromosome and unplaced sequences. Also for the buffalo, no indications have been reported on Chr Y and, so far, only the studies by FISH allowed correctly placing five genes on the MSY (Table 1, Figure 1) [40,41,43–45, 47,48].
The buffalo PAR is located on the telomere of q-arm and contains seven genes already mapped (Table 2, Figure 2) [45, 39,52,53].
Ovis aries/Capra hircus
The assembly of OAR and CHI genomes are Oar_rambouillet_v1.0. and Goat CVASU_BBG_1.0, respectively. The presence and precise location of five genes on the MSY were confirmed by FISH (Table 1, Figure 1) [40,41,43–45,47,48].
The PAR is located on the telomere of p-arm and it contains twenty four and nineteen genes physical mapped respectively for OAR and CHI (Table 2, Figure 2) [45,39,52,53].
PIG Y-CHROMOSOME AND PSEUDOAUTOSOMAL REGION
The domestic pig, Sus scrofa (SSC), plays a key role in the meat industry, but it acquired great importance also as biomedical animal model for many human diseases. An updated version of its genome sequencing has been recently published as Sscrofa11.1. Despite the new version, the annotation is fully available for the autosomes and the X chromosome, while little data is available for the Chr Y. Thanks to the cytogenetic investigation by BAC clones and FISH [54–56], it was possible to obtain information about the Y morphology and the evolution of sex chromosome genes. The Chr Y is the smallest of the pig chromosomes, metacentric and constituted of about 50 Mb in length as assessed by flow cytometry [57,58]. Chr Y short arm (Yp) is characterized by the presence of the most male-specific single-copy genes (physical mapped). Furthermore, given the highly repetitive nature of the long arm, to date, only a single copy sequence (DYZ1) is mapped on this chromosome portion (Table 1, Figure 1).
Regarding the information of the porcine PAR region, of sixteen loci only one is located in the terminal short arms of the sex chromosomes. Currently, the loci already known are only those cytogenetically mapped [55]. Moreover, the PAB lies next the shroom family member 2 (SHROOM2) or most proximal gene (Figure 2).
HORSE Y-CHROMOSOME AND PSEUDOAUTOSOMAL REGION
The horse, Equus caballus (ECA), is an economically and culturally important domestic species. The equine genome is approximately 2.68 Gb long and there are currently ~1,150 loci mapped by FISH with an average of one marker per ~2.5 Mb of the genome [59]. The ECA Y-chromosome is a small submetacentric, and forty-six genes have been mapped, as shown in Table 1 and Figure 1 [60–62,24]. The ECA genome is derived from a female, so the Chr Y has been poorly characterized.
Subsequently, through the sequencing of cDNA libraries, the gene content and the complete map of the euchromatic region were defined [24]. The map covers both the PAR and MSY regions [60,63]. A total of 129 markers, 110 sequence-tagged site (STS) and 19 genes, were found in the PAR. This region includes the PAB, which is located between PRKXY and EIF1AY in the Y chromosome [64]. Studies conducted on ECA have shown the presence of duplicate genes both in MSY and PAR [63,65]. So far, this condition has been observed only in horses and, having no other information, further studies would be necessary to confirm or exclude these duplications also in other equids/perrisodactyls or mammals. To date, the current map contains approximately ~400 BAC clones [66]. In Paria’s study of the 2009, through the use of the direct cDNA selection method, 29 genes and expressed sequence tag (ESTs) were identified and 23 out of them were known to be specific to the horse Y chromosome. To date, a total of 37 genes/transcripts from the horse MSY region were identified and showed that 20 genes are X-degenerate with known orthologs in other Eutherian species. The remaining 17 genes were acquired or novel and identified so far only in the horse or donkey Y chromosomes [62]. The PAR region is located at the telomere of the short p-arm of the X chromosome and at the telomere of the long q-arm of the Y chromosome and contains 18 physical mapping genes (Table 2, Figure 2) [17].
ALPACA Y-CHROMOSOME AND PSEUDOAUTOSOMAL REGION
The alpaca, Vicugna pacos (VPA), is a species of Camelids originally from South America. The quality of its meat [67, 68], the opportunity to exploit it as a source of milk [69] and, especially, its fiber make this species of fundamental importance for the economy of many countries [70,71].
The reference genome for this species is the Vic.Pac 3.1, which has been annotated for about 90%, and identifies ~76% of the genome to chromosomes [72,73]. The cytogenetic map currently known for the alpaca consists of 281 genes representative of all chromosomes [72]. So far, no genes have been identified by cytogenetic mapping on the Y-chromosome, except for ten loci on the PAR Yq-ter region (Table 2; Figure 2) because the CHORI-246 BAC library derives from a female alpaca [74]. The Alpaca Y-chromosome, according to the most recent physical mapping (Jevit et al [75]) is a small submetacentric, the smallest among of Old-World camels [76], and the PAR region is located in the long arm.
DISCUSSION
In this work, we report on the physical Y-gene mapping by FISH in the main farm animals species. Considering the existing gap in the genome assembly for the Chr Y, the present review is of great importance as one of the first indications to consider for a correct assembly. Moreover, the markers mapped on the Chr Y may offer useful indications for selection and have pratical implication in breeding programs for the biological function they may carry out. In fact, genes located on the Y chromosome are essential for spermatogenesis and male fertility, as demonstrated, for example, in Holstein bulls by association studies between Y-linked gene copy number variations (PRAMEY, HSFY, and ZNF) and fertility traits [77,78].
Although Y chromosome has never been a direct target for selection, fertility is always a significant factor in determining livestock productivity. In the majority of breeding programs for the farm animals, including those treated in the present study, the female-to-male sex ratio is significantly higher than one for the combination of the intensive artificial selection and the use of artificial insemination technology with high breeding value males. This condition greatly reduces the number of blood lineages and increases the consanguinity, as well as, the inbreeding depression on productive and reproductive traits, including fertility. Therefore, addressing this gap of knowledge will also increase the potential use of Y Chr markers for breeding purposes.
Comparisons of genes mapped on Y-chromosome by FISH among farm animals
To date, using the FISH method, it has been possible to map physically 208 loci belonging to the Y-chromosome of the species reviewed in the present study. In particular, 89 loci mapped in the non-pseudoautosomal region (Table 1) and 119 in the PAR region (Table 2). Regarding the comparison between PAR regions there is an interesting observation to do. This region is in the distal part of the Y-chromosome for all the species studied, but in BTA/OAR/CHI/SSC it is at the telomere of the p-arm, while in ECA/BBU it was reported at the telomere of the q-arm (Figure 2). In addition, very recently, the same position has been confirmed also in VPA [75]. In particular, in this species, the Y-chromosome is very small and does not show distinct cytogenetic characteristic, so that it is difficult to identify the location of the centromere. Only with the use of the FISH technique by PAR BACs, it has been demonstrated that the alpaca Chr Y is submetacentric with a very small and short p-arm and the PAR located at the telomere in the long q-arm [75].
Seventy-five out of 119 loci mapped in the PAR region and reviewed in this study were reported in bovids, whereas the other 44 were mapped in the other species. More in details, seven loci were mapped exclusively in the bovids (ASMTL, IL3RA, NLGN4, DU171056, DXYS3, DXYS4, EST BE750429) with the ASMTL and DXYS3 identified in all the four species (BTA, BBU, OAR, and CHI). DXYS4 physically located only in BTA and BBU, IL3RA and NLGN4X present in all except BBU, DU171056 and EST BE750429 absents in CHI. Three loci were mapped exclusively in VPA (CLCN4, MID1, WWC3), whereas four were identified only in SSC (OBP, PUDP, SW949, SHOX). One locus, SHROOM2, was mapped in SSC and VPA, and six were exclusively identified in ECA (AKAP17A, ASMT, DHRSX, GTPBP6, PLCXD1, XG) (Table 2). PAR genes have maintained a high level of synteny and conservation between different species. Indeed, their comparison would lead to divide them into three sub-regions. The first consists of PPP2R3B, CRLF2, CSF2RA, IL3RA, SLC25A6, ASMTL, ZBED1, CD99, ARSD, ARSL, ARSH, ARSF, GYG2, MXRA5, PRKXY, DXYS4, and NLGN4 (sub-region 1), the second STS, PNPLA4, ANOS1, TBL1X and GPR143 (sub-region 2), the third presents only in bovids, with DU171056, DXYS3, EST BE750429.
Sub-region 1 is present in all the species we reviewed and the genes mapped in this area mostly maintained the same order. In fact, the sequences mapped by FISH in bovids and horses maintained synteny, with the exception of GYG2 in ECA, which is located between CD99 and ARSD and not between ARSF and MXRA5 as identified in bovids (Figure 2). Furthermore, in bovids, BTA and OAR-CHI showed the same genes mapped with the exception of CSF2RA present only in OAR, while in BBU, it is possible to find only three genes mapped out of the genes belonging to this subregion. In pig and alpaca, the order of this sub-region is reversed compared to other species, although only some of the sequences in this area have been mapped for both species (Figure 2). Furthermore, within the same sub-region 1 the order of MXRA5 and ARSF for pig, ARSF and CFS2RA for alpaca do not retain synteny compared to other species.
A similar matter is evident also for the sub-region 2, present in almost all the species of interest except for the ECA. In BTA and OAR-CHI the mapped sequences are the same. Among these, only TBL1X was not mapped to CHI and is, instead, the only gene present in BBU. For both pigs and alpacas this area appears in order before sub-region 1 (Figure 2). Furthermore, the STS gene in VPA mapped to the end of this area and not to the beginning as it can be found in BTA, OAR and SSC. Regarding the absence of this sub-region in the ECA, Janečka et al [24] reported a transposition of some PAR genes in the MSY region, such as TBL1Y, ANOS1Y, STSP1 (Figure 1). This transposition would facilitate the recombination between PAR and MSY. This situation would justify the deletions in the MSY region that can be found in horses with sexual development disorders [24].
About the genes of this subregion, TBL1X is implicated in the testosterone concentration, spermatogenesis, and sperm motility; it was used as marker and it resulted important for the predicition of reproductive performance [79].
The sub-region 3, as mentioned above, regards only bovids and it conserved in BTA, BBU, OAR, and CHI a perfect synteny.
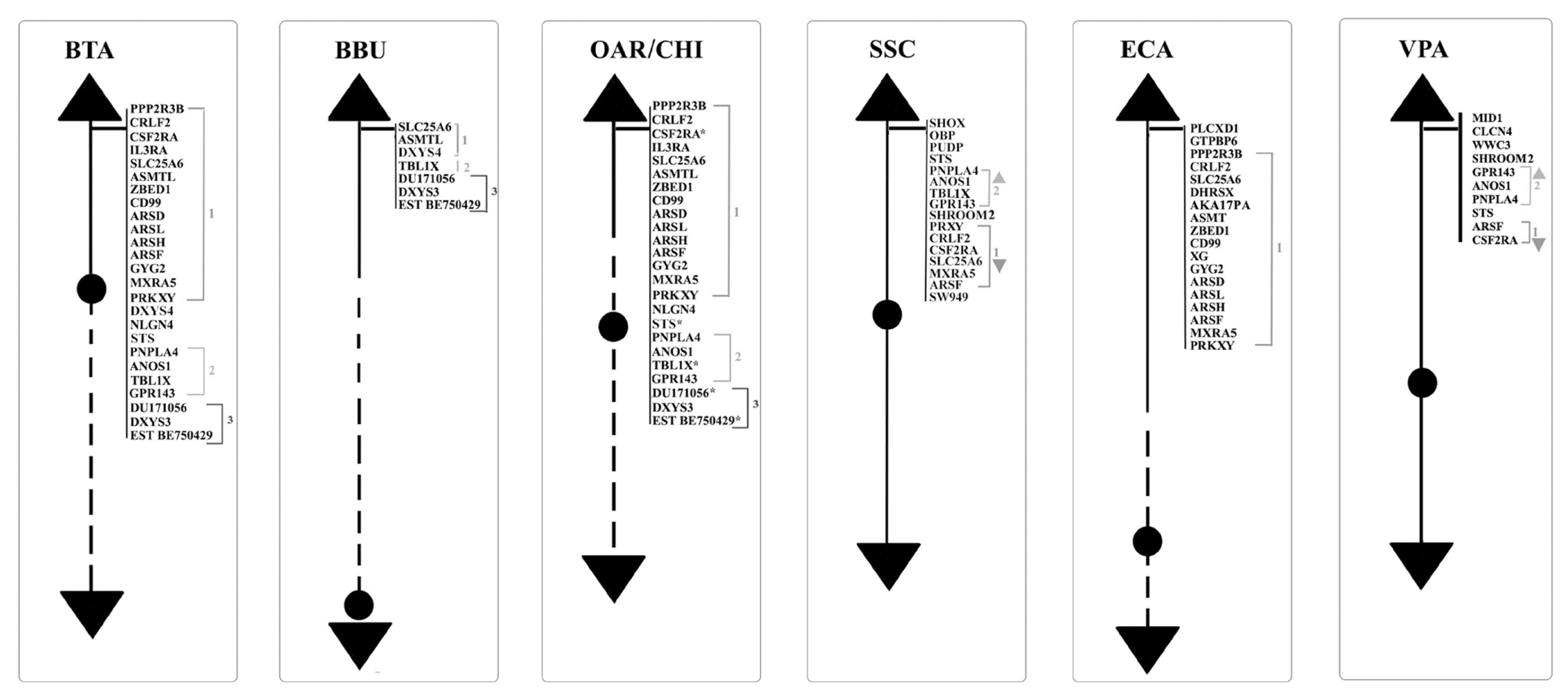
Concerning the 89 loci FISH mapped in the non pseudoautosomal region of the Chr Y (MSY), ECA represents the species reporting most of the loci currently mapped on Chr Y. Alone, it shows 46 positioned loci compared to BTA that has 18 mapped loci. About the other species, 10 loci were reported for SSC and 5 for BBU, OAR, and CHI. No genes were physically mapped to the alpaca Y-chromosome (Figure 1). Moreover, in this region, only the loci SRY, ZFY and TSPY1 were identified in most of the species investigated, that is BTA, OAR/CHI, SSC, and ECA; while BBU conserved only SRY and ZFY (Figure 3). The last two loci maintained a good synteny among the species, including in BBU where ZFY has been found in the telomeric region. In fact, according to Di Meo et al [45], the BBU Chr Y underwent a pericentric inversion compared to same chromosome in BTA. A similar situation has been described also for UMN0504 that is located near the PAR region only in these two species [45]. Regarding the locus TSPY1, it preserved synteny in the distal area of the short arm [48,42] in BTA, OAR and CHI. Conversely in SSC, this gene has been always reported near the centromeric region of the short arm [54]; and in ECA it has been reported in the euchromatic region of Y q-arm [60].
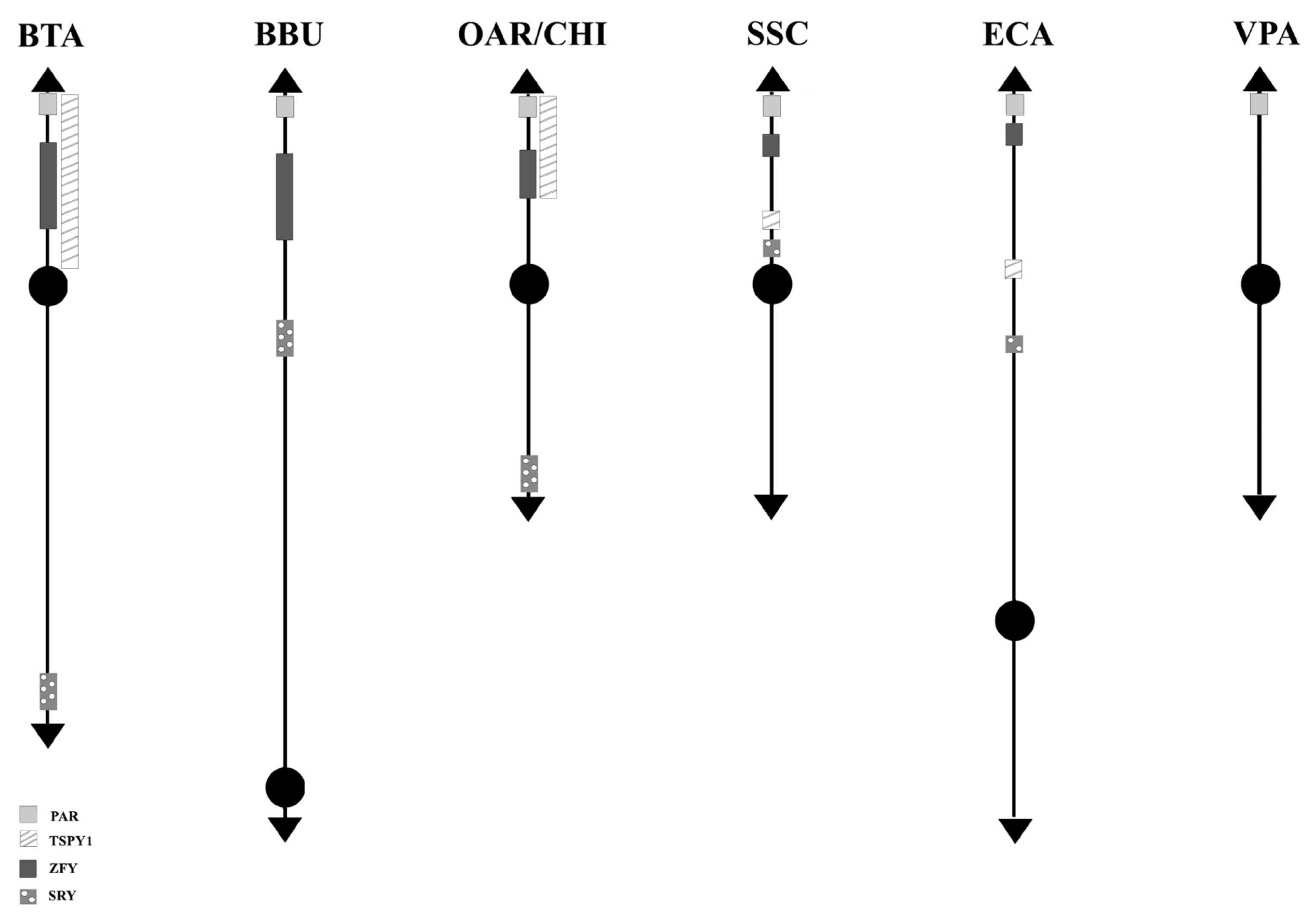
Schematic representation of SRY-ZFY-TSPY1 loci along the Y-chromosomes. No information has been found in VPA so far. VPA, Vicugna pacos.
SRY, ZFY, and TSPY1 have been considered very important and used as markers for sexual screening as they are involved in spermatogenesis and sexual differentiation [80–82].
DYZ1, a male-specific repeat DNA sequence, has been mapped only in BTA and SSC Y [54]. In particular, concerning BTA, the gene maps in the Yp13-q12 region with a higher concentration in the centromeric region [40], as confirmed by Habermann et al [46].
The AMELY, EIF2S3, USP9Y, DDX3Y, UBA1, and UTY genes were mapped only in SSC and ECA.). In SSC, AMELY is the first gene after the PAR region. However, according to Quilter et al [54], the orientation of the loci changes in ECA as a result of two rearrangements, so that the genes have the following order: EIF2S3, AMELY, USP9Y, DDX3Y, UTY, and UBA1 (Figure 1) [60,62]. Among them, AMELY is very important in the selection breeding programs because it represents a marker for the sex diagnosis of abnormalities involving the Y chromosome [83].
Instead EIF2S3, DDX3Y, UTY of this region together with SHROMM2 and SRY of MSY region were investigated for the level of their expression in the amniotic fluid [84]. This information can be very important in early sex determination by ensuring a targeted breeding program in species of economic interest.
UMN0304 and DYZ10 were mapped only in bovids. The former locus in BTA and OAR/CHI maps to the proximal pericentromeric regions of the p- and q-arms, whereas in BBU covers almost the entire Y chromosome except for the R-positive telomeric band. The latter locus maps in BTA and BBU as a painting, and in OAR/CHI to the proximal pericentromeric region of the p- and q-arms [45]. As regards DYZ10 and its different mapping, it must be considered that the data in literature are outdated in times former than the advent of new molecular technologies that allowed genomic libraries to be screened selectively. It would be interesting to localize again this gene, as well as many others, with new technologies like the PacBio sequencing [85] to be associated with classical methods such as FISH or Fiber-FISH. Concering the latter method, the Fiber-FISH is a technique that allows a significantly higher mapping resolution due to the direct visualization of chromatin fibers released from interphase nuclei that are extremely less condensed than the metaphase chromosomes observed by FISH. The Fiber-FISH is often used for determining size-gapping problems, locating delections, resolving chromatin breakpoints linked to diseases, estimating gene copy number variations, orientations, genes length, etc. [86,87].
Of the remaining genes, twelve were mapped only in cattle, and 37 in horses, as reported in Figure 1. The great interest in this species can be traced back to its ancient origins. The horse is in fact one of the first domesticated species that is integrated in the human working life as well as in his leisure activities [88]. Over the years, the interest in this species has increased because it is used in many rehabilitative activities or because it is used as a reference for the study of many pathologies, also considering the strong synteny between the chromosomes of this species with those of humans [89]. In addition, the fertility of stallions plays an essential role in the equine breeding industry (especially for racehorses), and although fertility is of primary importance for all species of zootechnical interest, this information is often limited also in this species.
From these results, the FISH technique turns out to be still a powerful tool contributing to the reduction of gaps currently present in sequencing processes.
The sequencing processes, in fact, in particular related to the Y-chromosome for livestock species, are still very complex and, in many respects, still little known due to the difficulty of assembling heterogametic genomes and the presence of highly repetitive ampliconic regions [15]. In addition, the sequencing of heterogametic genomes requires greater depth than in the homogametic genomes, resulting in an increase in costs [90]. One of the methods of sequencing sex chromosomes is based on the alignment of the heterogametic genome reads with the homogametic ones of the same species. In this way, those regions that are little reproduced on the homogametic chromosomes, are highlighted and are specific of the heterogametic chromosome [91].
As regards the PAR region, this Y-chromosome part is evolving [92,93], but, so far, only a few reports of PAR comparisons between species other than humans and mice have been undertaken [65,39].
Implementing comparative studies on animal genomes by FISH physical mapping of markers along the chromosomes, it would allow a better understanding of the evolutionary process in the different species, including the discovery of complex rearrangements [45] and the filling up of the current gaps in animal genomes.
Furthermore, the knowledge of a correct Y-genes assembly along the chromosome will give the opportunity to investigate more intensively their epistasis effects [94] for instance in the regulation of autosomal gene expression or in the control of the individual fitness. In fact, as demonstrated in humans [95] and Drosophila [96] the Y-chromosome carries multiple genes that differentially affect the expression of hundreds of X-linked and autosomal genes with a functional impact on microtubule stability, metabolism and spermatogenesis [97]. Such epistatic genetic effect might be considered a consequence of the autosomal nature of both sex chromosomes [98], so that prior to becoming sex chromosomes have been involved in autosome-autosome interactions. Thus, although the Y chromosome is a specialized part of the genome, it can play a key role on autosomal gene expression and individual fitness by interacting with the rest of the genome [99].
CONCLUSION
In the recent years, the genome of many species has been completed, including farm animals of economic interest. However, portions, mostly concerning the Y chromosome, are still unknown or not correctly assembled. The difficulty lies in the absence of reference points that can validate sequence data generated by Next-Generation Sequencing (NGS). In this respect, a real contribution to reduce the gap of knowledge and possible assembly errors on Chr Y can come from the use of the FISH combined with its derived techniques like the Fiber-FISH. The Fiber-FISH would allow to walk on all chromosome starting from specific markers in order to cover any gap, establish the correct order of very near genes or identify the right location of old and new repeated sequences.
Furthermore, the physical map by FISH could be of support also in the study of the cytological characteristics of the chromatin that controls the gene expression and regulation when combined with immunoassay techniques.
The data reported in this mini-review may be a starting point for further studies and future applications in animal science.
ACKNOWLEDGMENTS
The authors wish to thank Mr. Raffaele Pappalardo (CNR-ISPAAM) for the technical support.
Notes
CONFLICT OF INTEREST
We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.
FUNDING
This study was funded by RIS Bufala, project number PAUA_RIC_N_COMP_21_01.