Relationship among porcine lncRNA TCONS_00010987, miR-323, and leptin receptor based on dual luciferase reporter gene assays and expression patterns
Article information
Abstract
Objective
Considering the physiological and clinical importance of leptin receptor (LEPR) in regulating obesity and the fact that porcine LEPR expression is not known to be controlled by lncRNAs and miRNAs, we aim to characterize this gene as a potential target of SSC-miR-323 and the lncRNA TCONS_00010987.
Methods
Bioinformatics analyses revealed that lncRNA TCONS_00010987 and LEPR have SSC-miR-323-binding sites and that LEPR might be a target of lncRNA TCONS_00010987 based on cis prediction. Wild-type and mutant TCONS_00010987-target sequence fragments and wild-type and mutant LEPR 3′-UTR fragments were generated and cloned into pmiR-RB-REPORTTM-Control vectors to construct respective recombinant plasmids. HEK293T cells were co-transfected with the SSC-miR-323 mimics or a negative control with constructs harboring the corresponding binding sites and relative luciferase activities were determined. Tissue expression patterns of lncRNA TCONS_00010987, SSC-miR-323, and LEPR in Anqing six-end-white (AQ, the obese breed) and Large White (LW, the lean breed) pigs were detected by real-time quantitative polymerase chain reaction; backfat expression of LEPR protein was detected by western blotting.
Results
Target gene fragments were successfully cloned, and the four recombinant vectors were constructed. Compared to the negative control, SSC-miR-323 mimics significantly inhibited luciferase activity from the wild-type TCONS_00010987-target sequence and wild-type LEPR-3′-UTR (p<0.01 for both) but not from the mutant TCONS_00010987-target sequence and mutant LEPR-3′-UTR (p>0.05 for both). Backfat expression levels of TCONS_ 00010987 and LEPR in AQ pigs were significantly higher than those in LW pigs (p<0.01), whereas levels of SSC-miR-323 in AQ pigs were significantly lower than those in LW pigs (p<0.05). LEPR protein levels in the backfat tissues of AQ pigs were markedly higher than those in LW pigs (p<0.01).
Conclusion
LEPR is a potential target of SSC-miR-323, and TCONS_00010987 might act as a sponge for SSC-miR-323 to regulate LEPR expression.
INTRODUCTION
Obesity is a major source of human morbidity and mortality and is becoming more common worldwide [1]. Excess fat accumulation is a contributing factor to severe human diseases such as type II diabetes, cancers, and cardiovascular disease [2]. Therefore, it is important to understand the detailed mechanisms controlling adipogenesis and energy homeostasis. Currently, animal models for obesity research mainly are pigs, mice, and rats [3–5]. The pig is emerging rapidly as a biomedical model of energy metabolism and obesity in humans because it is postnatally devoid of brown fat and because of similar metabolic features, cardiovascular systems, and proportional organ sizes [6].
The products of the leptin receptor (LEPR) gene belong to the class I cytokine receptor family and were first reported in mice [7]. Poor expression of LEPR in certain tissues was suggested to lead to leptin resistance, which is commonly associated with obesity [8]. Ban et al [9] concluded that the knockdown of LEPR affects the mRNA expression of its downstream genes, suggesting that chicken LEPR plays a certain role in regulating the complicated gene network of preadipocytes. In pigs, a non-synonymous exonic polymorphism in the LEPR gene has been reported to be strongly associated with fat content in Iberian×Landrace and in Duroc×Landrace/Large White crossbreds [10]. The porcine LEPR gene is located between the 3rd subband and the 5th subband of the long arm 3 region of chromosome 6 and is located at the quantitative trait locus that is related to backfat thickness [11]. Compared to that of pigs, the nucleic acid homology of human and mouse LEPR sequences are 89.2% and 80.3%, and the amino acid sequence alignment is 86.5% and 76.6%, respectively [12]. LEPR has thus been considered a genetic marker associated with body composition, growth rate, and obesity in some pig breeds [13].
LncRNAs are greater than 200 nt in length and have been implicated in diverse biological processes such as cell-cycle regulation, genomic imprinting, and cell differentiation [14]. Transcriptome profiling revealed that lncRNAs are expressed at a lower level than protein-coding transcripts and are more developmental stage- and tissue-specific [15]. Studies have confirmed that lncRNAs play a role in adipogenesis. For example, ADINR RNA (adipogenic differentiation induced noncoding RNA, a lncRNA that is transcribed from a position about 450 bp upstream of the CCAAT/enhancer binding protein α [C/EBPα] gene) specifically binds PA1 (PAX-Interacting protein 1 [PAXIP1]-associated glutamate rich protein 1 [PAGR1], also known as PA1) and recruits MLL3/4 histone methyltransferase complexes to increase H3K4me3 and decrease H3K27me3 histone modifications at the C/EBPα locus, resulting in gene activation and enhanced adipogenesis [16]. Further, the PU.1 antisense lncRNA binds and negatively regulates the level of PU.1 mRNA, suppresses protein expression, and thus improving PU.1-mediated peroxisome proliferator-activated receptor γ (PPARγ) expression [17].
miRNAs are small non-coding RNAs that can function widely by modulating multiple physiological processes such as development, immunity, cell differentiation, apoptosis, metabolism, and signal transduction [18]. These molecules also regulate fat growth, mainly by acting on transcription factors that regulate the signaling pathways involved in adipocyte differentiation or by preventing cell proliferation to promote or inhibit the differentiation of fat cells [19]. Studies have demonstrated that lncRNAs can interact with protein-coding genes through direct competition for miRNA binding [20]. For example, Liu et al [21] found that lncRNA Gm15290 sponges miR-27b to promote PPARγ-mediated adipogenesis in vitro and increase fat deposition and body weight in high fat diet-fed mice.
Based on previous high-throughput sequencing studies by our study group, we identified some differentially-expressed lncRNAs in the backfat tissues of Anqing six-end-white (AQ, the obese breed) and Large White (LW, the lean breed) pigs. Among them, we were interested in lncRNA TCONS_ 00010987, because the cis mechanism prediction indicated that its potential target gene was LEPR. Considering the physiological and clinical importance of this gene in regulating obesity and the fact that porcine LEPR expression has not been previously shown to be under lncRNA control, we conducted a bioinformatic analysis to predict miRNAs that could bind SSC-TCONS_00010987-target sequence fragments and the SSC-LEPR-3′-UTR. We found that SSC-miR-323 could directly bind lncRNA TCONS_00010987 and LEPR mRNA with low free energy of binding. Next, we constructed dual luciferase reporter gene vector with an SSC-TCONS_00010987 target fragment or an SSC-LEPR-3′-UTR, as well as similar vectors with mutations in the respective SSC-miR-323-binding sites. The dual luciferase assay system was used to detect relative luciferase activities. Additionally, the backfat tissue expression patterns of lncRNA TCONS_00010987, SSC-miR- 323, and LEPR in LW and AQ pigs were investigated. This study aimed to provide a theoretical basis for the possible mechanism through which SSC-miR-323 and lncRNA TCONS_ 00010987 regulate LEPR expression.
MATERIALS AND METHODS
Ethics statement
All animal experiments were conducted according to the Regulations and Guidelines for Experimental Animals established by the Ministry of Science and Technology (Beijing, China, revised in 2004) and approved by the Institutional Animal Care and Use Committee of Anhui Agricultural University (Permit number: SYXK 2016-007).
Animals and tissue sampling
Six purebred castrated male AQ pigs and six purebred castrated male LW pigs (with a body weight of approximately 100 kg) were used in this work. Strictly fast for 24 hours before slaughter, free to drink water. The heart, liver, spleen, lung, kidney, longissimus dorsi muscles, and backfat tissues were rapidly harvested from the carcasses and immediately frozen in liquid nitrogen, and all tissue samples were stored at −80°C prior to RNA extraction. The another longissimus dorsi muscles were collected to test the content of intramuscular fat.
Measurements of carcass traits
The backfat thicknesses (mm), intramuscular fat of the longissimus dorsi muscles (%), carcass lean meat percentage (%) and loin eye area (cm2) of AQ and LW pigs were measured strictly in accordance with the Rules for performance determination of breeding pigs (NY/T 822–2004) issued by the ministry of agriculture of the People’s Republic of China. Intramuscular fat content was determined by measuring the crude fat of muscle using Soxhlet Extraction with petroleum ether.
Quantitative real-time reverse transcription polymerase chain reaction
Total RNA from the heart, liver, spleen, lung, kidney, longissimus dorsi muscles, and backfat tissues were extracted using the RNAeasy Mini Kit (Qiagen, Hilden, Germany), following the operating manuals. Total RNA was checked for purity and quantity with a Nanodrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA). RNA integrity was measured by denaturing agarose electrophoresis and a Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA).
For quantitative analysis of lncRNA TCONS_00010987 and LEPR mRNA expression, 1 μg of total RNA from each sample was reverse-transcribed into cDNA using the Prime Script RT Kit (TaKaRa, Tokyo, Japan) at 37°C for 15 min and 85°C for 5 s in a 10-μL reaction mixture, according to the manufacturer’s instructions. The glyceraldehyde-3-phosphate dehydrogenase gene was used as an endogenous control gene.
For the quantitative analysis of SSC-miR-323 expression, 1 μg of total RNA from each sample was reverse-transcribed into cDNA using the NCode EXPRESS SYBR GreenER miRNA qPCR Kit (Invitrogen, Carlsbad, CA, USA) at 37°C for 1 h in a 20-μL reaction mixture, according to the manufacturer’s instructions. Porcine U6 small nuclear RNA (U6) was used as an endogenous control gene.
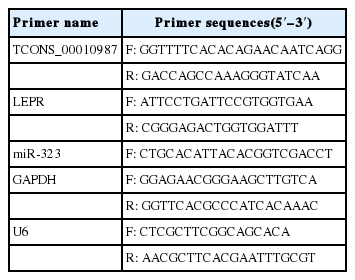
All primers used are listed in Table 1. Quantitative polymerase chain reaction (qPCR) was performed using the CFX96 TouchTM Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). Thermal cycling conditions consisted of 95°C for 30 s, followed by 40 cycles at 95°C for 5 s and 60°C for 30 s. Relative lncRNA TCONS_00010987, LEPR, and SSC-miR-323 expression levels were calculated from Ct values using the 2−ΔΔCT method. All tests were carried out in triplicate.
Bioinformatics analysis
The porcine LncRNA TCONS_00010987 sequence was obtained from our previous high-throughput sequencing results. The cis prediction of target genes was based on the lncRNAs regulating their neighboring or nearby genes. The 10-kb upstream or downstream region of lncRNA TCONS_00010987 was searched for its target gene. According to cis prediction, LEPR was selected as a potential mRNA target of lncRNA TCONS_00010987 for follow-up studies.
The SSC-miR-323 sequence was obtained from the miRDB website (http://www.mirdb.org/mirdb/policy.html) and miRNA–mRNA interactions were obtained from three reliable online miRNA-target databases as follows: BiBiServ (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/), miRDB (http://www.mirdb.org/mirdb/policy.html), and RNA22 v2 (https://cm.jefferson.edu/rna22/Interactive/). miRNA-lncRNA interactions were predicted using BiBiServ. Based on the predictive criteria including having good complementarity with targeted sequences, being bound to targeted sequences with low free energy of binding, and being conserved among species, SSC-miR-323 was selected as a candidate miRNA that interacts with TCONS_00010987 and LEPR for follow-up studies.
Plasmid construction
Based on the porcine lncRNA TCONS_00010987-target sequence, a partial segment containing the SSC-miR-323-bound sequence and a mutated segment of the lncRNA TCONS_ 00010987-target sequence in which the SSC-miR-323-bound sequence TAATGTG was converted to ATTACAC were constructed by biosynthesis (Supplementary Figure S1). The expected length of both amplicons was 950 bp. The biosynthesized fragments were inserted between the XhoI and NotI sites of the pmiR-RB-REPORTTM firefly luciferase reporter vector (RiboBio, Guangzhou, China). Recombinants harboring the desired biosynthesis products were confirmed by DNA sequencing. The recombinant plasmids were termed “WT” (pmiR-RB-REPORTTM-SSC-TCONS_00010987-wild-type target sequence fragments) and “MT” (pmiR-RB–REPORTTM-SSC-TCONS_00010987-mutant target sequence fragments).
Based on the porcine LEPR mRNA 3′-UTR sequence (NM_ 001024587.1), a partial segment containing the SSC-miR-323-bound sequence and a mutated segment in which the SSC-miR-323-bound sequence AATGTG was converted to TTACAC were constructed by biosynthesis (Supplementary Figure S2). The expected length of both amplicons was 766 bp. The biosynthesis products were then subcloned into the pmiR-RB-REPORTTM firefly luciferase reporter vector (RiboBio, China) between XhoI and NotI sites. Recombinants harboring the desired biosynthesis products were confirmed by DNA sequencing. The recombinant plasmids were termed “WT” (pmiR-RB-REPORTTM-SSC-LEPR-wild-3′-UTR) and “MT” (pmiR-RB-REPROTTM-SSC-LEPR-mutant-3′UTR).
SSC-miR-323 mimics and the negative control were purchased from RIBOBIO (RiboBio, China).
Plasmid transfection and luciferase assays
For luciferase assays to test the interaction between SSC-TCONS_00010987 and SSC-miR-323, HEK293T cells were transiently co-transfected with 100 ng of pmiR-RB-REPORTTM-SSC-TCONS_00010987-WT or pmiR-RB-REPORTTM-SSC-TCONS_00010987-MT and 100 nM of SSC-miR-323 mimics or its negative control using Lipofectamine 3000 (Invitrogen, USA) in 96-well plates.
For luciferase assays to test the interaction between SSC-LEPR and SSC-miR-323, HEK293T cells were transiently co-transfected with 100 ng of pmiR-RB-REPORTTM-SSC-LEPR-WT or pmiR-RB-REPORTTM-SSC-LEPR-MT and 100 nM SSC-miR-323 mimics or its negative control using Lipofectamine 3000 (Invitrogen, USA) in 96-well plates.
Forty-eight hours after transfection, the luciferase activities were determined using the Dual-luciferase Reporter Assay System according to the manufacturer’s instructions (Promega, Madison, WI, USA). All reactions were performed in triplicate.
Protein extraction and western blotting
Total proteins were extracted from backfat using radio immunoprecipitation assay lysis buffer (Beyotime, Shanghai, China) and protein content was measured using the bicinchoninic acid protein assay kit (Beyotime, China). Total protein (5 to 20 μL) was loaded onto a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel, separated by electrophoresis, and transferred onto a polyvinylidene difluoride membrane. Blots were blocked with 5% skim milk overnight at 4°C and blotted with specific primary antibodies for LEPR (1:300, bs-20498R, Bioss, Beijing, China) and β-actin (1:2,000, TA-09, ZSGB-BIO, Beijing, China). The bound primary antibodies were determined with a horseradish peroxidase conjugated secondary antibody goat anti-rabbit immunoglobulin G (IgG) (1:2,000, ZB2301, Zsbio, Beijing, China) and a horseradish peroxidase conjugated secondary antibody goat anti-mouse IgG (1:2,000, ZB2305, Zsbio, China), respectively. The data were analyzed using Image J software (GEL system, Beijing kcrx bio-company, Beijing, China). The relative expression of LEPR was calculated as the ration of optical density of LEPR to that of β-actin.
Statistical analysis
The values are expressed as the mean±standard deviation. A Student’s t-test was used to evaluate the statistical differences between two groups and p<0.05 was considered significant, whereas p<0.01 was considered highly significant.
RESULTS
Carcass traits of Anqing Six-end-white and Large White pigs
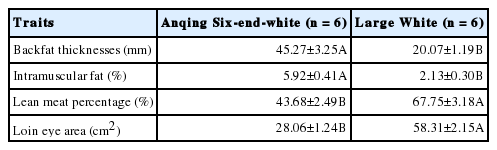
As shown in Table 2, the average backfat thicknesses of AQ and LW pigs were 45.27±3.25 and 20.07±1.19 mm, respectively. The loin eye area of LW pigs is 107.80% higher than that of AQ pigs (p<0.01), and the lean meat percentage is 55.11% higher than that of AQ pigs (p<0.01). The intramuscular fat of AQ pigs is 177.93% higher than that of LW pigs (p<0.01). Based on the extreme differences in their fat phenotypes, these two breeds comprised good models for our study.
LncRNA TCONS_00010987 interacts with SSC-miR-323
Based on bioinformatics analyses, we identified one putative binding site between lncRNA TCONS_00010987 and SSC-miR-323 (Figure 1a). Gel electrophoresis results showed that the biosynthesis products containing the corresponding target sequence fragments were specific and of the expected size (Figure 1b). The sequencing results of vector containing the SSC-TCONS_00010987-wild-type target sequence fragments are shown in Figure 1c; the fragment had 100% sequence identity with the corresponding region of the porcine TCONS_ 00010987 gene obtained by our previous high-throughput sequencing results. Sequencing results of the vector harboring the SSC-TCONS_00010987-mutant target sequence fragments are shown in Figure 1d; as shown, the TAATGTG sequence was successfully mutated to ATTACAC, without changes to other bases. These results confirmed that TCONS_00010987-wild-type target sequence fragments and TCONS_00010987-mutant target sequence fragments were successfully cloned into the dual luciferase reporter vectors.

LncRNA TCONS_00010987 is a putative target of SSC-miR-323. (a) Representation the lncRNA TCONS_00010987- and miR-323-binding sites by BIBISERV. (b) Identification of PCR fragments by agarose gel electrophoresis. M, DNA marker; lane 1, PCR fragment of the lncRNA TCONS_00010987 target sequence; lane 2, PCR fragment of the lncRNA TCONS_00010987 mutant target sequence. (c) Sequencing results of the dual luciferase reporter vector containing the lncRNA TCONS_00010987 target sequence. (d) Sequencing results of the dual luciferase reporter vector containing the lncRNA TCONS_00010987 mutant target sequence. PCR, polymerase chain reaction.
As shown in Figure 2, compared to expression with the negative control, SSC-miR-323 mimics significantly inhibited luciferase activity from the wild-type SSC-TCONS_00010987 target sequence fragments (p<0.01); however, SSC-miR-323 mimics had no effect on luciferase activity when the SSC-miR-323-binding site was mutated. These results indicated that miR-323 targets TCONS_00010987 via the SSC-miR-323- binding site located in the TCONS_00010987-target sequence fragments and inhibits its transcriptional activity.
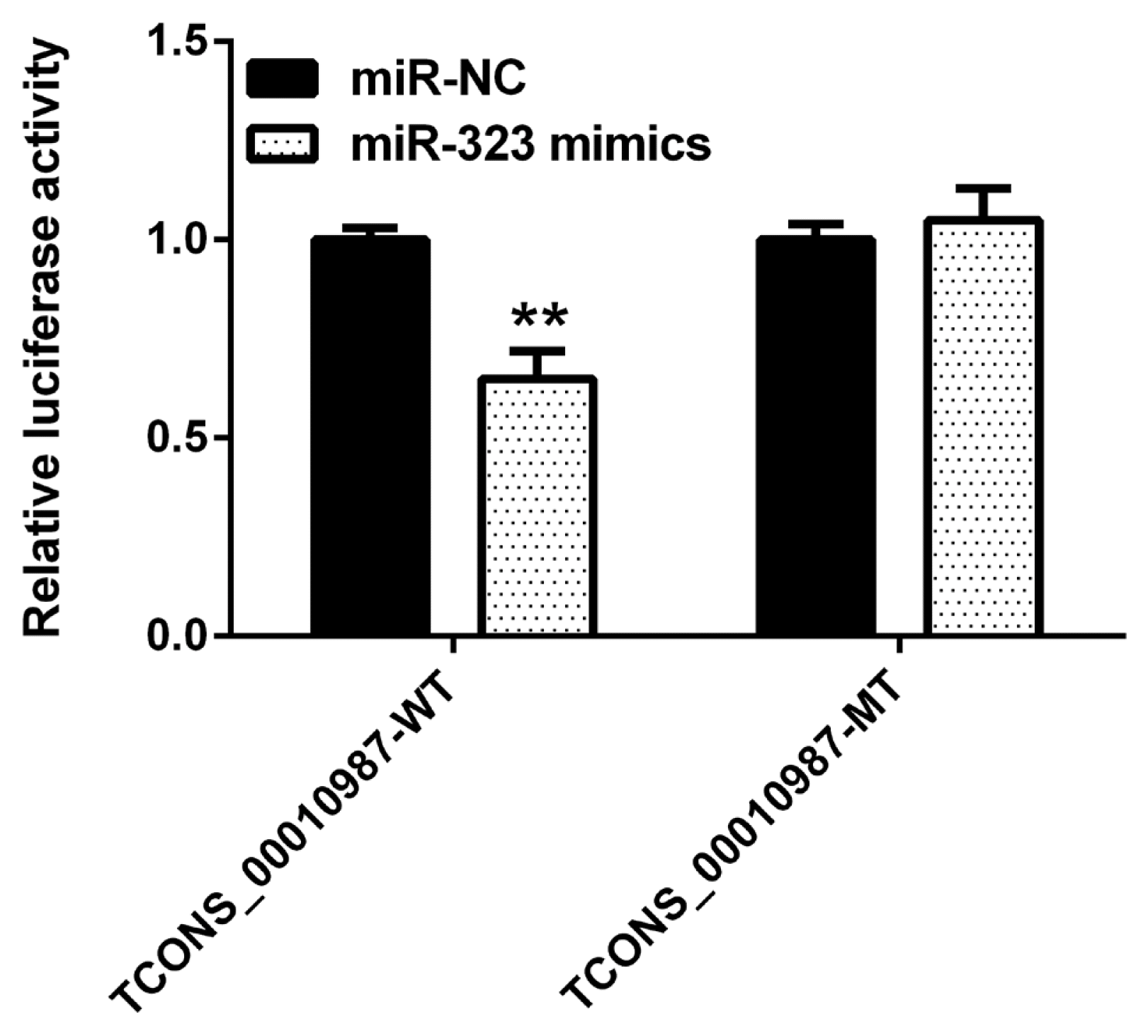
Luciferase activity in HEK293T cells transfected with reporter vectors containing lncRNA TCONS_00010987 target sequence fragment variants and SSC-miR-323 mimics. WT, wild-type TCONS_00010987 target sequence; MT, TCONS_00010987 target sequence with a mutation in the SSC-miR-323-binding site. Data are presented as the mean±standard deviation of n = 3 experiments. ** p<0.01.
Leptin receptor is directly targeted by SSC-miR-323
One putative binding site between LEPR and SSC-miR-323 was identified using bioinformatic analysis (Figure 3a). Gel electrophoresis results showed that the biosynthesis products containing the corresponding 3′-UTR were specific and of the expected size (Figure 3b). Sequencing results of the SSC-LEPR-wild-type-3′-UTR vector are shown in Figure 3c, whereas those of the SSC-LEPR-mutant-3′-UTR vector are shown in Figure 3d; here, the AATGTG sequence was successfully mutated to TTACAC, without changes to other bases. These results confirmed that the LEPR-wild-type-3′-UTR and LEPR-mutant-3′-UTR were successfully cloned into the dual luciferase reporter vectors.

LEPR is a putative target of SSC-miR-323. (a) Diagram of the putative binding sites between LEPR and SSC-miR-323 based on bioinformatics prediction. (b) Identification of PCR fragments by agarose gel electrophoresis. M, DNA marker; lane 1, PCR fragment of LEPR mRNA 3′-UTR sequence; lane 2, PCR fragment of LEPR mRNA 3′-UTR mutant sequence. (c) Sequencing results of the dual luciferase reporter vector containing the LEPR 3′-UTR. (d) Sequencing results of the dual luciferase reporter vector containing the LEPR 3′-UTR with a target sequence mutation. LEPR, leptin receptor; PCR, polymerase chain reaction.
As shown in Figure 4, compared to that with the negative control, SSC-miR-323 mimics significantly inhibited wild-type SSC-LEPR 3′-UTR activity (p<0.01). However, these mimics had no effect on luciferase activity from the SSC-LEPR mRNA 3′-UTR reporter containing the mutated SSC-miR-323-binding site. These results indicated that SSC-miR-323 targets LEPR via the miR-323-binding site located in the 3′-UTR of the LEPR mRNA to inhibit its transcriptional activity.
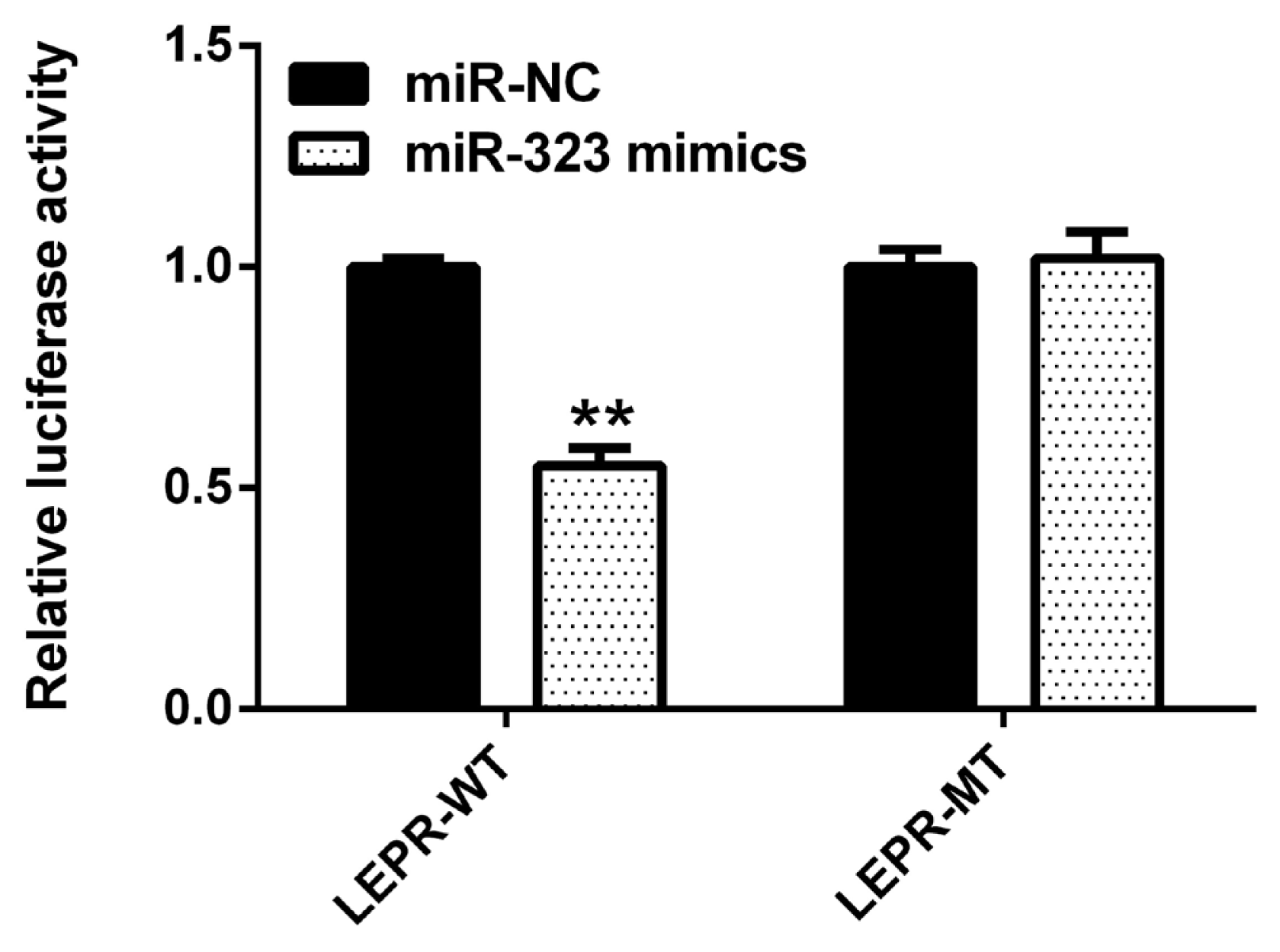
Luciferase activity in HEK293T cells transfected with reporter vectors containing LEPR mRNA 3′-UTR variants and SSC-miR-323 mimics. WT, wild-type LEPR mRNA 3′-UTR; MT, LEPR mRNA 3′-UTR with a target sequence mutation in the SSC-miR-323-binding site. LEPR, leptin receptor. Data are presented as the mean±standard deviation of n = 3 experiments. ** p<0.01.
Backfat expression patterns of TCONS_00010987, miR-323, and LEPR in Anqing Six-end-white and Large White pigs
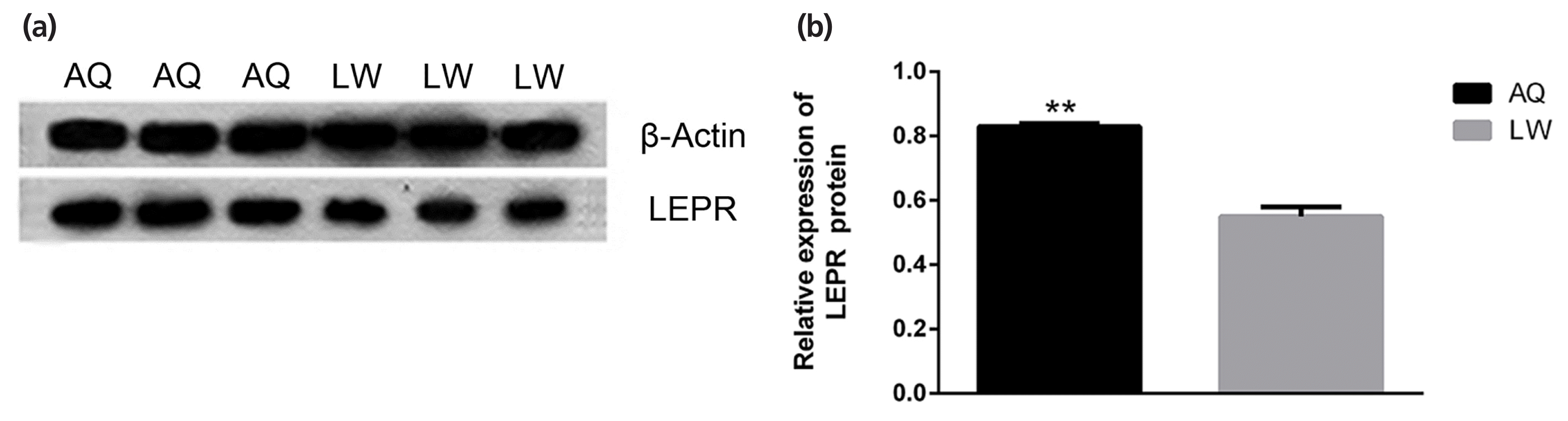
As shown in Figure 5, qPCR analysis revealed that lncRNA TCONS_00010987, LEPR, and SSC-miR-323 were all highly expressed in porcine backfat tissues. Furthermore, expression levels of lncRNA TCONS_00010987 and LEPR in AQ pigs were significantly higher than those in LW pigs (p<0.01), whereas levels of SSC-miR-323 in AQ pigs were significantly lower than those in LW pigs (p<0.01). As shown in Figure 6, western blotting results showed that LEPR protein levels in the backfat tissues of AQ pigs were also markedly higher than those in LW pigs (p<0.01). As indicated, the backfat expression levels of lncRNA TCONS_00010987, LEPR mRNA, and LEPR protein in obese pigs were significantly higher than those in the lean breed, whereas levels of SSC-miR-323 showed the opposite expression trends.
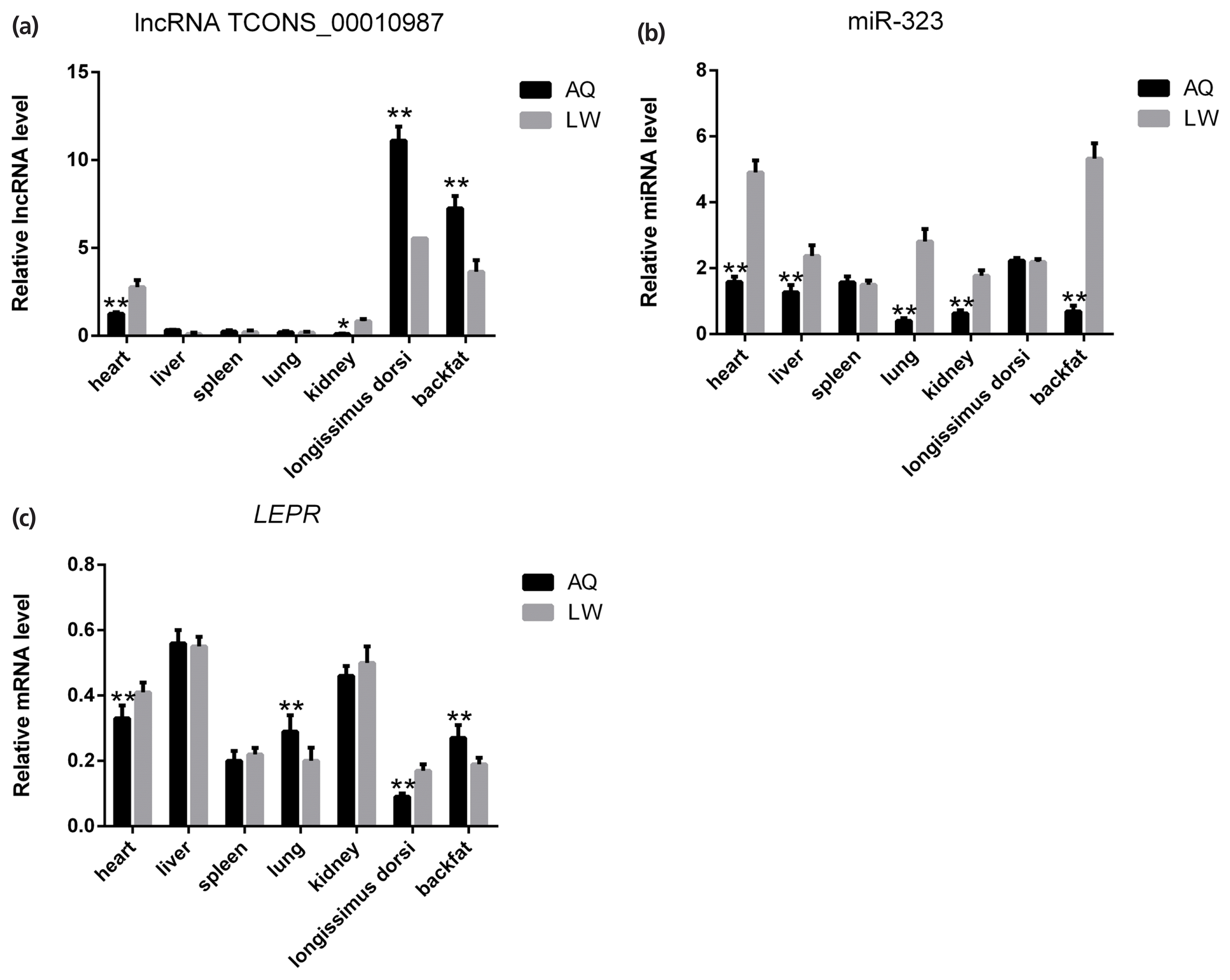
Differential expression of lncRNA TCONS_00010987, miR-323, and target genes in tissues of Anqing Six-end-white and Large White pigs. (a) lncRNA TCONS_00010987; (b) miR-323; (c) LEPR mRNA. LEPR, leptin receptor. Data are presented as the mean±standard deviation (n = 6). * p<0.05, ** p<0.01.
DISCUSSION
In this study, we used LW pigs and AQ pigs to comprise lean-fat models based on their significant phenotype differences in carcass fat deposition traits (as shown in Table 2). Lean pig breeds, such as Large White (LW), have been intensively selected over the past decades for improved growth rate and muscularity [22]. AQ pigs, one Chinese fat-type pig breed, have high good meat quality and slow growth rate [23]. Whereas the Chinese domestic breed has the universal characteristics of higher fat deposition capacity compared to that with Western commercial pig breeds [24]. Accordingly, the two pig breeds can be taken as ideal animal models to comparatively study the mechanisms controlling adipogenesis.
Leptin receptors interact with leptin to play a wide range of roles in animals, such as controlling the body mass balance, regulating body fat content and regulating feed intake [25]. Adipocytes synthesize and secrete leptin, which in turn acts on the hypothalamus to maintain the stability of body fat [8]. At high levels, leptin inhibits food intake through the melanocortin receptor system, and at low levels, it promotes food intake through neuropeptide Y. Mutations in the LEPR gene result in leptin insensitivity, hyperphagia, morbid obesity, and metabolic and endocrine abnormalities [26]. In pigs, LEPR mRNA was expressed in adipose tissue, brain, muscle, fat, liver, hypothalamus, pituitary gland and reproductive tissues (endometrium and ovary) [27]. Studies have confirmed that the expression of LEPR in the muscle and fat tissues in high-fat pigs is significantly higher than that in low-fat pigs [28,29]. Based on our results, we confirmed this; specifically, LEPR protein and mRNA expression levels were both significantly higher in the backfat of obese breed pigs (AQ) than in that of lean breed pigs (LW). It was speculated that the increase of LEPR expression was caused by increased fat deposition, which promoted the decomposition of fat and controlled the body mass balance in pigs.
Su et al [30] showed that SSC-miR-323 is significantly increased in the myocardial tissue of pigs with coronary microembolization. Further, Skovgaard et al [31] found that miR-323 might be involved in controlling acute influenza infection in pigs. However, to date, there had been virtually no research on miR-323 in pig fat. Based on our results, SSC-miR-323 was expressed in the backfat tissues of both AQ and LW pigs; however, the expression patterns in obese and lean pigs differed, indicating that SSC-miR-323 might be associated with fat metabolism in pigs.
LncRNA TCONS_00010987 is a novel lncRNA, for which the sequence was obtained based on our previous high-throughput sequencing; moreover, the coding potential calculator analysis suggested coding potential to have a score <0 [32], demonstrating that it is a non-coding RNA. The results of this experiment showed that lncRNA TCONS_00010987 expression levels in the backfat differ greatly between the AQ and LW pigs. Specifically, we demonstrated that whereas lncRNA TCONS_00010987 is indeed expressed in this tissue, expression levels differ depending on whether the animal is obese or lean.
Increasing studies have reported that many lncRNAs function as competing endogenous RNAs (ceRNAs) by serving as sponges that bind and sequester miRNAs. For example, Lv et al [32] revealed that lncRNA Unigene56159 promotes epithelial–mesenchymal transition by acting as a ceRNA for miR-140-5p in hepatocellular carcinoma cells. Further, Zhao et al [33] provided evidence that lncRNA-plasmacytoma variant translocation 1 functions as an endogenous ‘sponge’ by competing for miR-448 binding to regulate expression of the target SERBP1, thereby promoting the proliferation and migration of pancreatic cancer cells. Finally, Shang et al. found that lncRNA TCONS_00041960 enhances osteogenesis and inhibits adipogenesis in rat bone marrow mesenchymal stem cells by targeting miR-204-5p and miR-125a-3p [34].
In the present study, bioinformatics analyses predicted a link among porcine lncRNA TCONS_00010987, SSC-miR-323, and LEPR. Luciferase-reporter assays confirmed that lncRNA TCONS_00010987 might act as a sponge for SSC-miR-323 to regulate LEPR expression. In addition, qPCR and western blot analysis indicated that the backfat expression patterns of lncRNA TCONS_00010987, LEPR mRNA, and LEPR protein in obese and lean pigs were opposite to those of SSC-miR-323.
In conclusion, luciferase activity and expression pattern assays indicated that lncRNA TCONS_00010987 might act as a sponge for SSC-miR-323 to regulate LEPR expression. These results, suggesting that SSC-miR-323 can bind LEPR and TCONS_00010987, indicate that further study is required to fully elucidate the role of SSC-miR-323, TCONS_00010987, and LEPR in the epigenetic mechanisms that affect fat metablism in pigs. These results have implications in understanding the detailed mechanisms controlling adipogenesis, energy homeostasis, and obesity.
Supplementary Data
ACKNOWLEDGMENTS
This work was supported by the Natural Science Foundation of Anhui Province (1508085QC52), the planning subject of ‘the twelfth five-year-plan’ in national science and technology for the rural development in China (2015BAD03B01), the key subject of study and research of Anhui province (1704a 07020085, 1704a07020090), the Open project supported by State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences (GREKF15-03), the National Natural Science Foundation of China (31972531), Anhui Provincial Science and Technology Major Project (17030701008, 17030701061), and the Anhui Provincial Modern Industrial Technology System of Swine.
Notes
CONFLICT OF INTEREST
We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript. Gao Yafei is an employee of Anhui Haoxiang Agriculture and Animal Husbandry Co. LTD.