Variance Component Quantitative Trait Locus Analysis for Body Weight Traits in Purebred Korean Native Chicken
Article information
Abstract
Quantitative trait locus (QTL) is a particular region of the genome containing one or more genes associated with economically important quantitative traits. This study was conducted to identify QTL regions for body weight and growth traits in purebred Korean native chicken (KNC). F1 samples (n = 595) were genotyped using 127 microsatellite markers and 8 single nucleotide polymorphisms that covered 2,616.1 centi Morgan (cM) of map length for 26 autosomal linkage groups. Body weight traits were measured every 2 weeks from hatch to 20 weeks of age. Weight of half carcass was also collected together with growth rate. A multipoint variance component linkage approach was used to identify QTLs for the body weight traits. Two significant QTLs for growth were identified on chicken chromosome 3 (GGA3) for growth 16 to18 weeks (logarithm of the odds [LOD] = 3.24, Nominal p value = 0.0001) and GGA4 for growth 6 to 8 weeks (LOD = 2.88, Nominal p value = 0.0003). Additionally, one significant QTL and three suggestive QTLs were detected for body weight traits in KNC; significant QTL for body weight at 4 weeks (LOD = 2.52, nominal p value = 0.0007) and suggestive QTL for 8 weeks (LOD = 1.96, Nominal p value = 0.0027) were detected on GGA4; QTLs were also detected for two different body weight traits: body weight at 16 weeks on GGA3 and body weight at 18 weeks on GGA19. Additionally, two suggestive QTLs for carcass weight were detected at 0 and 70 cM on GGA19. In conclusion, the current study identified several significant and suggestive QTLs that affect growth related traits in a unique resource pedigree in purebred KNC. This information will contribute to improving the body weight traits in native chicken breeds, especially for the Asian native chicken breeds.
INTRODUCTION
Quantitative trait loci (QTLs) are genomic regions associated with quantitative traits. To identify these regions analyses should focus on identifying QTLs that link phenotypic data (quantitative traits) and genotypic data (usually molecular markers) in an attempt to explain the genetic basis of variation in complex traits (Andersson and Georges, 2004). The DNA markers usually used for QTL detection are single nucleotide polymorphisms (SNPs), restriction fragment length polymorphisms (RFLPs), and simple sequence length polymorphisms (SSLPs or microsatellites) (Vignal et al., 2002). For the past several decades, microsatellite markers have dominated animal genome studies, especially in the field of molecular genetics, due to the high allele number at a single microsatellite locus and the possibility to genotype by simple polymerase chain reaction (PCR). The high numbers of alleles allow generation of high values of heterozygosity where the number of reference families can be dramatically reduced to construct the map. Utilization of microsatellite markers has been widely used to map the genome in agricultural animals including chicken, which has made the chicken a valuable organism in genomics studies (Groenen et al., 2000). In addition, microsatellite markers have been valuable in detecting QTL regions for economically important traits in the chicken (Zhang et al., 2011; Wang et al., 2012).
In Korea, genetic improvement of Korean native chicken (KNC) has been a major concern since 1994, when the government established a conservation program dedicated to KNC restoration. This project has been successfully divided the KNC into five lines based on plumage colors consisting of black, gray, red, white and yellow lines (Jin et al., 2014). One of these lines has been used to create a commercial three-way KNC hybrid designated Woorimatdag. Consumer acceptance of KNC has steadily increased due to their superior nutritional contents, taste and unique texture of meat (Jeon et al., 2010). Also, the price of native chicken is approximately twice that of commercial broilers, which has allowed poultry breeders to develop a native chicken industry in Korea (Cahyadi et al., 2013; Seo et al., 2013). However, KNC have some inferior performance (growth-related) traits. As in most native chicken breeds, KNC displays low growth rate and high variation in body weight traits. Therefore, appropriate breeding strategies are required to improve the growth performances of KNC to meet consumer’s demands, so the proportion of imported chicken breeds in the market can be reduced (Cho et al., 2014).
In the present study, microsatellite markers were used to genotype chicken samples and genome-scan analysis was conducted to map QTLs for body weight traits of KNC. Mapping of QTLs using microsatellite markers has been successfully used to determine genetic contributions to economically important traits in chickens by examining the relationship between variations of microsatellite allelic and growth-related traits (Schreiweis et al., 2005; Tercic et al., 2009). Mapping of QTL regions will not be clearly valuable without screening and tracing the positional candidate genes in those QTL regions. For this reason, QTL studies are always followed by detection of mutations in the functional regions within QTL regions. Uemoto et al. (2010) identified a positional candidate gene (the ornithine decarboxylase gene) for growth and carcass weight (CW) in F2 intercross chickens based on a previously determined QTL region on chicken chromosome 3 (GGA3). The retinoblastoma 1 gene in the QTL region for body and CW is also a potential positional candidate gene for body weight (Zhang et al., 2011).
Hence, the present study was performed to map QTLs for body weight traits using variance component linkage analyses and to trace positional candidate genes within those QTL regions in KNC.
MATERIALS AND METHODS
Experimental animals
The experimental design used in the present study was F1 nuclear family design comprising two generations (i.e., F0 and F1). A total of 88 F0 that comprises 15 sires and 68 dams has been mated by following within line mating system. Total of 3 sires and 4 to 5 dams for each line have been used to produce 595 F1 progeny (black, 90; gray, 110; red, 135; white, 125; and yellow, 135). These animals were generated under very strict breeding procedure of the National Institute of Animal Sciences (NIAS), Korea. Dams were mated to sires by artificial insemination and matings between sibs were avoided. Additionally, they were maintained and reared under standard feeding and environmental conditions.
Genotypic and phenotypic analyses
The DNA genome was isolated by following the DNA extraction procedure for blood samples (GeNet Bio, Daejeon, Korea). NanoDrop 2000c UV-Vis Spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA) was used to quantify and to check the quality of extracted DNA. Then, the DNA samples were stored in the refrigerator at −20°C to maintain DNA quality for further analyses. Total of 131 highly polymorphic microsatellite (MS) markers and 8 SNP markers were used to genotype F0 and F1 samples. A total of 27 chromosomes (26 autosomes+Z chromosome) have been covered with the total map length of 2,729.4 centi Morgan (cM) and the average distance between markers was 19.64 cM. These MS markers were selected from the Ark Database (http://www.thearkdb.org/arkdb/) to be genotyped that previously described by Seo et al. (2013). In addition, 8 SNP markers were selected from our candidate genes SNPs data that generated by direct sequencing and genotyped using PCR-RFLP and Fluidigm Genotyping Technology. Detailed information on genetic linkage map was reported by Seo et al. (2015b). Four markers on Z chromosome covering 113.3 cM were not used for QTL analysis; i.e., a total of 135 DNA markers covering 26 autosomes were used in this study.
Phenotypic measured in current study was body weight and growth rate traits. Body weights were observed and recorded at every two weeks from hatch to slaughter age at 20 weeks of age. Weight of half carcass was also measured when slaughter process and evisceration were totally completed.
Quantitative trait loci mapping
The QTL analysis was performed using a variance component based program, SOLAR, for outbred nuclear F1 resource pedigree (Almasy and Blangero, 2010). The identical-by-decent (IBD) matrix was calculated based on the marker and pedigree information. Using this IBD matrix, the logarithm of the odds (LOD) score to test presence of QTL was computed at a 1-cM interval across the genome for each F1 chicken. The IBD matrix is incorporated to a linear mixed model as follows:
where, y is a vector of the phenotypic values for body weight traits except for the hatch weight; b is a vector of fixed effects including sex, line, and batch; q is a vector of additive QTL effect; u is a vector of residual additive polygenic effects; e is a vector of residual environmental effects; X, Z1, and Z2 are incidence matrices for b, q, and u, respectively. The mean and variance for residual additive polygenic effects can be defined as: u~N (0, Aσa2), where A is the additive genetic relationship matrix computed from the F1 pedigree in this study and σa2 is the residual additive polygenic variance. The mean and variance for additive QTL effects can be defined as: q~N (0, Gσq2), where G is the IBD matrix and σq2 is QTL effect variance. The mean and variance for residual effects can be defined as: e~N (0, Iσe2), where I is the identity matrix and σe2 is residual variance. The variance components were estimated by maximum likelihood method.
A likelihood ratio test using SOLAR was conducted to evaluate the significance of the full model (with random common environmental effects of dam) compared to the reduced model (without random common environmental effects of dam). As a result, the Model 1 was extended further to incorporate the maternal (dam) environmental effect for the hatch weight trait as follows:
where, y is a vector of the phenotypic values for the hatch weight; m is a vector of random maternal (dam) environmental effects; Z3 is incidence matrix for m. The mean and variance for the maternal (dam) effects can be defined as: m~N (0, Sσm2), where S is the structuring matrix for the maternal (dam) environmental effects and σm2 is variance due to dam.
Thresholds for evaluating significance of QTL were calculated by a numerical method (Piepho, 2001). The threshold levels established by this method vary as a function of the trait studied and the chromosome length. A 1% chromosome-wide threshold for the significant linkage was employed. A suggestive linkage was employed using a 5% chromosome-wide threshold. The 1-LOD drop method was used to estimate the support intervals (SIs) for the identified QTL (Lander and Botstein, 1989).
Screening of positional candidate genes
Positional candidate genes for growth-related traits in the identified QTL region were screened using National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov), Ensamble Genome Browser (http://www.ensembl.org). The relevance of all screened positional candidate genes to growth phenotypes was verified using the GeneCards database (http://www.genecards.org). Moreover, SNP markers from the whole genome sequence of KNC (Seo et al., 2015a) were assigned to the selected positional candidate genes. We further screened the candidate genes that included heterozygous SNPs.
RESULTS
Phenotypic analysis
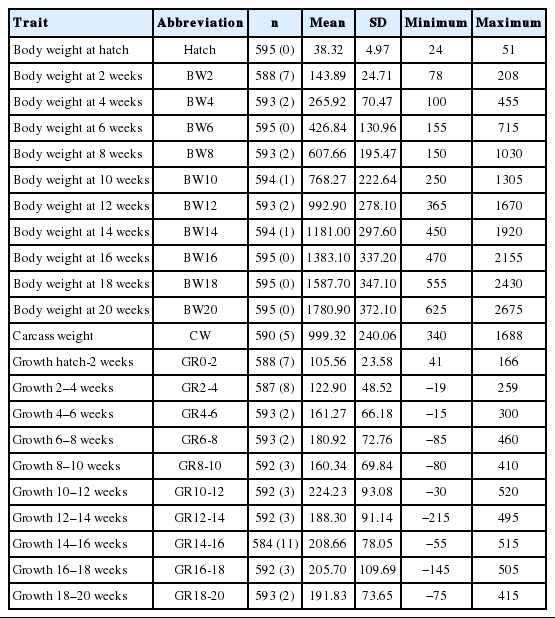
The descriptive statistics of phenotypic traits in F1 population including mean, standard deviation, minimum and maximum values are presented in Table 1. The number of chickens ranged from 584 to 595. The Ryan-Joiner normality evaluation was also performed to ensure all observed data were in normal distribution. To evaluate the significance of random common maternal (dam) environmental effects, a likelihood ratio test was conducted. These maternal effects were significant only for hatch weight (p<0.001). Thus, Model 2 was used for the QTL analysis of hatch weight. Model 1 was used to perform QTL mapping of all other traits, since the dam effects were not significant for other body weight traits (data not shown).
Quantitative trait loci analysis
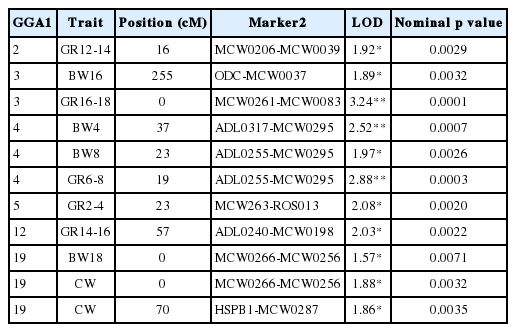
QTLs that reached suggestive thresholds for body weight, half CW, and growth are summarized in Table 2. Five QTLs influencing growth traits were identified in this F1 resource pedigree of KNC. Among them, two QTLs identified at the 1% chromosome-wide significance levels: GGA 3 for growth 16–18 weeks (GR16-18) (LOD = 3.24) (Figure 1A) and GGA4 for GR6-8 (LOD = 2.88) (Figure 1B). Rest of them were mapped on GGA2, GGA5, and GGA12. Four additional QTLs affecting body weight traits were detected at the 1% (i.e., QTL on GGA4 for BW4) and 5% (i.e., QTLs on GGA3, GGA4, GGA19) chromosome-wide threshold levels. The QTL for body weight at 16 weeks of age (BW16) was identified on GGA3. Similar patterns of QTL curves were found on GGA4 that affected body weight at 4 weeks of age (BW4) and 8 weeks of age (BW8) (Figure 1B). These QTLs were located around 23 to 37 cM on GGA4, where the closest marker to the QTL peak was ADL0203. The patterns of these QTL peaks similarly appeared at early to middle stages of age, and gradually disappeared when body weights were measured at 16 to 20 weeks of age (data not shown). In addition, a QTL for body weight at 18 weeks of age (BW18) was detected on GGA19 between the MCW0266 and MCW0256 markers.

Test statistic profiles for body weight and growth traits on GGA3 (A), GGA4 (B), and QTL for body weight (BW) 18 and (carcass weight) CW on GGA19 (C). The y-axis represents the LOD score testing the hypothesis of a QTL in a given genomic position. Marker map with distances between markers in Kosambi centi Morgan (cM) is given on the x-axis. Colored lines represent QTL profiles for each trait. The horizontal thick and thin black lines represent the 1% and 5% chromosome-wide significant thresholds, respectively. GGA, chicken chromosome; QTL, quantitative trait loci; LOD, logarithm of the odds; cM, centi Morgan.
Regarding CW, two suggestive linkage QTLs at the 5% chromosome-wide threshold level were also identified on flanking regions of GGA19 located at 0 and 70 cM. One of these QTLs was in a similar location as the QTL for BW18 (Figure 1C).
Positional candidate genes
The candidate genes were intensively traced just after QTL for the body weight and growth were detected. The screening of the positional candidate genes was carried out on GGA4 since these QTL signal consistently appeared for BW4, BW8 and GR6-8 (Figure 1B). The QTL affecting GR6-8 (LOD = 2.88, nominal p value = 0.0003) was used to screen for a positional candidate gene. The QTL confidence interval was established by the 1-LOD drop method. A positional candidate gene was located in the QTL region for GR6-8 on GGA4. The candidate was the insulin receptor substrate 4 gene (IRS4) that plays an important role in growth, reproduction and glucose homeostasis (Sadagurski et al., 2014).
DISCUSSION
Two significant (GGA3 and GGA4) and three suggestive (GGA2, GGA5, and GGA12) QTLs for growth were identified. One significant and three suggestive QTLs affecting body weight traits were also detected on three different chromosomes. They comprised a QTL for BW16 on GGA3, two neighboring QTLs located at 23 to 37 cM on GGA4 for BW4 and BW8, and a QTL for BW18 on GGA19. Moreover, two QTLs for CW were also identified at two distal ends of GGA19.
The QTL for GR16-18 on GGA3 showed the highest LOD score (LOD = 3.24, nominal p-value = 0.0001) in this study. Interestingly, similar QTL linkage patterns for BW4, BW8 and GR6-8 on GGA4 were detected (Figure 1B). In fact, there were several non-significant QTLs that showed very similar QTL curve patterns compared to the three identified QTLs (data not shown). The LOD score of those QTLs on GGA4 were also decreased by age and gradually disappeared after 16 weeks of age (data not shown). Therefore, the same QTL (i.e. pleotropic QTL) may affect these traits. However, we cannot exclude the possibility that two closely linked but different QTLs may also affect these two traits. These results indicate that the QTL detected on GGA4 may affect growth traits from early to middle life stages, and those the QTLs on other chromosomes affect body weight traits at different stages of age. Thus, chicken growth and development may involve multiple genes that have roles at different stages of life.
Studies have been conducted to detect QTL for body weights in different chicken populations (Wahlberg et al., 2009; Wang et al., 2012; Sheng et al., 2013). Wang et al. (2012) identified two QTL locations for body weight at 2 to 5 weeks of age, and body weight 8 to 10 weeks of age on GGA3. Ikeobi et al. (2004) reported that GGA3 (where the QTL for BW16 was presently detected) could be a candidate QTL for breast muscle weight. A QTL on GGA19 was identified as an additional QTL affecting BW18 with the nearest marker to the QTL peak being MCW0256. This MS marker was analyzed as a single marker to be associated with body weight at 3 weeks of age in a chicken interline cross (Atzmon et al., 2008). QTL regions on GGA4 were frequently detected in early to middle life of KNC, even though the 5% significance chromosome-wide level was only found for BW4 and BW8. Thus, this QTL region has potential value for the screening of positional growth-related candidate genes, since other studies also reported that the same QTL regions were detected for BW4, BW6, BW8, BW10, and BW12 in different chicken populations (Jacobsson et al., 2005; Wahlberg et al., 2009; Podisi et al., 2013). Other QTL regions were also identified on GGA4 for overall body weight, CW wing weight, drum stick weight, and thigh weight (Ikeobi et al., 2004; Schreiweis et al., 2005; Tsudzuki et al., 2007). Therefore, along with GGA1, GGA4 is implicated as a chromosome that harbors QTLs for growth-related traits (Cahyadi et al., 2014). Regarding the QTLs for CW in KNC, two QTLs were located at 0 and 70 cM on GGA19. Liu et al. (2013) performed a genome-wide association study to identify loci for body composition and meat quality traits in Beijing-You chickens. A QTL for CW was detected on GGA19 located at 1.8 cM. This location is very close to the results of the current study (0 cM) where a QTL for CW was detected in the KNC purebred population. The presently detected QTL for CW located at 70 cM on GGA19 is a novel QTL for CW in the chicken. As for growth, QTL for GR16-18 on GGA3 was revealed as the strongest evidence for linkage in this study. Abasht and Lamont (2007) performed a genome-wide association study to identify loci for fatness in chicken. They could map a QTL for abdominal fat percentage to the similar genomic region of our QTL for GR16-18. Interestingly, significant QTL for GR6-8 was co-localized with QTLs for BW4 and BW8. This result indicated the genetic co-regulation of QTL for body weight and growth.
In this study, the genetic linkage analysis was performed using 135 DNA markers (i.e., 127 MS markers+8 SNP markers) that representing 26 autosomal linkage groups. We could not cover 13 chromosomes in the chicken. Therefore, the QTLs identified in the current study do not account for the whole QTL effects since our genome scan could not cover several micro-chromosomes. In addition, there is lack of coverage on markers in some macro-chromosomes. Thus, we cannot exclude the possibility that these un-covered genome region may harbor additional significant QTLs.
To validate the identified markers in this study, we will search for the positional candidate genes in the QTL regions flanked by the markers. In fact, we have whole genome sequencing data from the five lines of KNC to develop SNP markers in KNC chicken genome (Seo et al., 2015a). After obtaining SNP information of positional candidate genes from whole genome sequence data, we have plans to perform association studies between these SNP markers and growth traits using a low density customized SNP chip assay. We would like to include the SNP markers located within the positional candidate genes for the customized SNP chip. If a marker shows significant association with growth traits, this information can be useful for marker-assisted selection to improve genetic potential of growth traits in KNC.
The investigation of positional candidate genes in QTL regions on GGA4 was performed. All genes in this region were extracted using the Biomart option in Ensembl (www.ensembl.org) and directly screened considering previously published relevant information regarding their roles and functions. Finally, a potential candidate gene, the insulin receptor substrate 4 gene (IRS4), was selected after an extensive manual curation. IRS4 acts as an interface between multiple growth factor receptors possessing tyrosine kinase activity, such as insulin-like growth factor 1 receptor (IGF1R), insulin receptor, and fibroblast growth factor receptor 1 (FGFR1), and plays an important role in development, growth, reproduction, and metabolic homeostasis (Hinsby et al., 2004). In addition, Sadagurski et al. (2014) revealed that IRS4 is involved in severe obesity and energy expenditure in mice. Based on the prior reports, IRS4 could be a promising novel candidate gene for body weight traits in the chicken. To confirm the effect of the IRS4 gene polymorphisms on body weight, an association study among genotypic and phenotypic should be performed.
Although previous studies have reported the efficiency in mapping QTL that accounts for genetic and phenotypic differences between two divergent lines, researches using the F2 intercross (or backcross) design have provided less practical information on whether these QTL present within the commercial population of interest. For the successful implementation of QTL information into selective breeding programs, segregation of QTL needs to be validated within the population of interest. Thus, the identified QTL regions in this study using F1 nuclear pedigree will provide practical information to identify molecular genetic factors that affect desired growth traits within the KNC population. Moreover, this information from this study will contribute to efforts to improve the body weight traits in native chicken breeds, especially Asian native chicken breeds.
ACKNOWLEDGMENTS
This study was funded by grants from the Next-Generation BioGreen 21 Program (Project No. PJ008133012014, Rural Development Administration), and the Golden Seed Project (No. 2013005042SB730, Korea Institute of Planning & Evaluation for Technology in Food, Agriculture Forestry & Fisheries), Korea.
Notes
CONFLICT OF INTEREST
We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.