Genetic Association of the Porcine C9 Complement Component with Hemolytic Complement Activity
Article information
Abstract
The complement system is a part of the natural immune regulation mechanism against invading pathogens. Complement activation from three different pathways (classical, lectin, and alternative) leads to the formation of C5-convertase, an enzyme for cleavage of C5 into C5a and C5b, followed by C6, C7, C8, and C9 in membrane attack complex. The C9 is the last complement component of the terminal lytic pathway, which plays an important role in lysis of the target cells depending on its self-polymerization to form transmembrane channels. To address the association of C9 with traits related to disease resistance, the complete porcine C9 cDNA was comparatively sequenced to detect single nucleotide polymorphisms (SNPs) in pigs of the breeds Hampshire (HS), Duroc (DU), Berlin miniature pig (BMP), German Landrace (LR), Pietrain (PIE), and Muong Khuong (Vietnamese potbelly pig). Genotyping was performed in 417 F2 animals of a resource population (DUMI: DU×BMP) that were vaccinated with Mycoplasma hyopneumoniae, Aujeszky diseases virus and porcine respiratory and reproductive syndrome virus at 6, 14 and 16 weeks of age, respectively. Two SNPs were detected within the third exon. One of them has an amino acid substitution. The European porcine breeds (LR and PIE) show higher allele frequency of these SNPs than Vietnamese porcine breed (MK). Association of the substitution SNP with hemolytic complement activity indicated statistically significant differences between genotypes in the classical pathway but not in the alternative pathway. The interactions between eight time points of measurement of complement activity before and after vaccinations and genotypes were significantly different. The difference in hemolytic complement activity in the both pathways depends on genotype, kind of vaccine, age and the interaction to the other complement components. These results promote the porcine C9 (pC9) as a candidate gene to improve general animal health in the future.
INTRODUCTION
The complement system is composed of more than 30 distinct humoral and cell surface proteins (Volanakis, 1998). Invading pathogens trigger this system as activation agents. As an enzyme, the prior proteins activate the subsequent proteins to generate C3-convertases, a key enzyme in the formation of membrane attack complex (MAC), which can lead to the formation of pores on the cell surface. Insertion of the complex through the cell membrane causes influx of extracellular material into the cell, loss of cytoplasm and therefore quickly results in death of the target cells. The MAC is a sequential self-assembly of one of each molecule of C5b, C6, C7, C8, and 6–18 molecules of C9 (Kolb et al., 1972; Podack et al., 1982; Tschopp et al., 1984; Muller-Eberhard, 1986). C9 is a single-chain protein that has the unique ability to polymerize during MAC formation (Tschopp, 1984; Volanakis, 1998). Although lysis of erythrocytes occurs even without C9 (Stolfi, 1968), its presence increases the rate of hemolysis (Hadding and Muller-Eberhard, 1969). In vertebrates, the C9 and perforin form oligomeric pores that lyse bacteria and kill virus-infected cells, respectively (Rosado et al., 2007). In pig, the pC9 was mapped to chromosome 16q14 using in situ hybridization method (Thomsen et al., 1998) and IMpRH tool (Do, 2010).
Kira et al. (1998) found C9 deficiency in 4 Japanese patients suffering from meningococcal meningitis. This deficiency was the result of a nonsense substitution in codon 95 in exon 4 of the C9 gene. The same mutation is cause of dermatomyositis and progressive inflammatory myopathy in a 28-year-old Japanese woman (Ichikawa et al., 2001). C9 deficiency was also described in a 29-year-old woman in good health but with low serum complement levels (CH50) (Inai et al., 1979). Hemolytic activity in classical pathway and alternative pathway was associated with variation of the porcine genes encoding the complement components pC3 (Wimmers et al., 2003), the pC5 (Kumar et al., 2004), and the porcine mannose-binding lectin 1 (pMBL1) and pMBL2 (Phatsara et al., 2007). In a recent study, Wimmers et al. (2011) promote the terminal complement components (pC6, pC7, pC8A, pC8B, and pC9) as strong candidates for disease resistance.
Here the pC9 gene was screened for polymorphism and examined for genetic association with hemolytic complement activity in both the classical and the alternative pathway.
MATERIALS AND METHODS
Experimental design
Experimental animals were the porcine breeds Hampshire (HS, n = 1), Duroc (DU, n = 1), Berlin miniature pig (BMP, n = 1), German Landrace (LR, n = 30), Pietrain (PIE, n = 30), and Muong Khuong (MK, Vietnamese potbelly pig, n = 25). These animals were used for identifying cDNA sequence, screening single nucleotide polymorphisms (SNPs), and genotyping.
A resource population of 417 F2 DUMI (Duroc×Berlin Minature Pig) animals, the product of backcross between DU and BMP (Hardge et al., 1999), was used to genotype the identified SNPs and study the association with hemolytic complement activity. Therefore, these animals were divided into two experimental groups i) unvaccinated control group (n = 36) and ii) trial group (n = 381) vaccinated with Mycoplasma hyopneumoniae (Mh, Tellamune, Mycoplasma, Pfizer, Karlsruhe, Germany), Aujeszky disease virus (ADV, Porcilis, Begonia Diluvac, Intervet, Tönisvorst, Germany), and porcine reproductive and respiratory syndrome virus (PRRS, Ingelvac PRRS MLV, Boehringer Ingelheim, Germany) at 6, 14, and 16 weeks of age, respectively. Ethylenediamine tetraacetic acid (EDTA) anticoagulated blood samples were taken before (day 0) and after Mh and ADV vaccination at day 4 and 10 but only at day 10 after PRRS vaccination (8 different sampling point times) and were immediately cooled on ice. Sera and plasma isolations were obtained within two hours by centrifugation at 4°C at a maximum speed of 14.000 rpm for 10 minutes and stored at −80°C for further immunological analysis. For DNA extraction and cDNA synthesis, tail, ear, and liver tissues were collected from adult animals, respectively. Just after slaughtering, liver was immediately frozen in liquid nitrogen or submerged in RNAlater buffer (Ambicon, Applied Biosystems, Darmstadt, Germany) according to the manufacturer’s instruction and stored at −70°C until further analysis.
Preparation of genetic materials
Preparation of genomic DNA from tail or ear samples was performed by standard proteinase K digestion followed by phenol-chloroform extraction and ethanol precipitation. Finally, the DNA was resuspended in 1×Tris-EDTA buffer and stored at 4°C (Do et al., 2010).
Total RNA was isolated from 100 mg liver tissue using the one step TRIzol Reagent (Chomczynski et al., 1993, 1995; Do et al., 2010) as recommended by the manufacturer (Sigma-Aldrich, Taufkirchen, Germany).
First-strand cDNA was synthesized from 1 μg of total RNA using 500 ng of oligo (dT)13 VN primer (Promega, Mannheim, Germany) and 500 ng random hexamer primers (Promega, Germany) (Do et al., 2010).
Primer design and amplicon verification
Based on GenBank entries (accession no. BP139003.1, CJ007506.1, BX920671, CF363669) the specific oligonucleotide primers, which could amplify the whole complete coding region of the pC9 cDNA sequence were designed using Primer3 tool (Rozen and Skaletsky, 2000). The primer sequences and their position in cDNA sequence of candidate genes are given in Table 1. Also these primer pairs were used to detect SNPs within the pC9 gene by comparative sequencing.
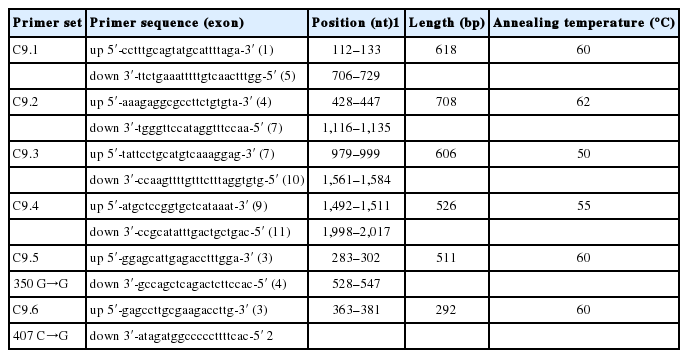
Oligonucleotide primer pairs used for sequencing, screening single nucleotide polymorphisms and genotyping
Polymerase chain reaction (PCR) was performed in 20 μL total volume containing 50 ng of liver cDNA or 100 ng of genomic DNA, 0.2 mM of each primer (forward or reverse primer), 50 μM of each dNTP (Roth, Karlsruhe, Germany), 0.5 U of Taq polymerase (Sigma-Aldrich, Germany) in 1×Taq buffer and, 1.5 mM of MgCl2. The PCR thermal cycling program was set up with an initial denaturation step of 94°C for 4 min followed by 40 amplification cycles (denaturation at 94°C for 30 s, annealing at 60°C for 30 s, elongation at 72°C for 1 min) and terminated by an extension at 72°C for 5 min using a T1 Research Thermocycler (Biometra, Göttingen, Germany). The PCR product was evaluated on 1% agarose Tris-acetate-EDTA gel stained with ethidium bromide for evaluation and documentation. Before sequencing by using ABI 310 Gene Analyzer (Applied Biosystems, Germany) or MegaBACE 1000 (GE Healthcare, München, Germany), the PCR product was purified by ethanol precipitation.
Detection of SNPs and genotype analysis
One animal of each of porcine breed HS, LR, DU, PIE, BMP, and MK was used for screening SNPs by comparative sequencing. The SNP sites were reconfirmed by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) genotyping of LR, PIE, and MK animals using the restriction enzymes BsrDI and HpyCH4III.
Study of association with hemolytic complement activity
To distinguish between the classical (CH50) and the alternative-pathway (AH50), the standard CH50 and AH50 hemolytic assays were applied according to the methods of Liu and Young’s (1988) and Demey et al. (1993). Briefly, hemolytic complement activity in the classical and alternative pathway were expressed as the amount of serum that caused a 50% hemolysis of antibody sensitised sheep red blood cells (SRBCs) (CH50, U/mL) and 50% hemolysis of rabbit erythrocytes (AH50, U/mL) in the reaction mixture, respectively (Wimmers et al., 2003). Accordingly, for classical complement reaction SRBCs were sensitised with hemolysin (anti-sheep red blood cell stroma from rabbit, Sigma, Taufkirchen, Germany) diluted 1:75 in Veronal buffered saline (VBS) buffer (containing 3.12 mM Diethylbarbituric acid; 1.82 mM Diethylbarbituric acid sodium salt; 145.45 mM NaCl; 1 mM MgCl2; 0.15 mM CaCl2; pH 7.45; conductivity 13.5 mS). Sensitised SRBCs were used as a 2% cell-suspension. For testing alternative complement pathway activity rabbit erythrocytes (RRBC) were used as foreign surfaces. The RRBC were resuspended with GVBS-Mg-EGTA buffer (containing 0.1% gelatin; 3.12 mM Diethylbarbituric acid; 1.82 mM Diethylbarbituric acid sodium salt; 145.45 mM NaCl; 1 mM MgCl2; 20 mM Ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid (EGTA); pH 7.45; conductivity 13.5 mS) to a final concentration 0.5%. Serial dilutions of test sera (50 μL) in veronal buffer (VBS or GVBS-Mg-EGTA, respectively) were made in duplicate in flat-bottomed 96-well microtitre plates (1:2, 1:4, 1:8…). 50 μL of sensitized SRBCs or RRBC suspension were added to each serum dilution, respectively. Plates were incubated for 90 min at 37°C. Absorbance at 650 mn was determined using a microplate reader. The readings were transformed by the method of von Krogh according to Mayer (1961) and the hemolytic titre was expressed as the titre that lysed 50% of the erythrocytes (CH50 U/mL, AH50 U/mL).
Statistical analysis
In this study, the SAS’s PROC MIXED procedure combined with REPEATED statement (The SAS software package, release 9.1) were used for analyzing the variance of experimental dataset to estimate the effect of genotypes of candidate genes on complement activity at eight different time points of vaccination. The analysis model address valid standard errors of the fixed-effect factor estimates in order to identify other significant environmental and genetic effects apart from the factor of genotypes and its interaction by stepwise elimination of non-significant effects.
Where yijklmno is the hemolytic complement activity (CH50 and AH50), μ is overall mean, sirei is the fixed-effect of sire (i = 1 to 3), damj is the fixed-effect of dam (j = 1 to 11), parityk is the fixed-effect of parity (k = 1 to 5), treatmentl is the fixed-effect of treatment-vaccinated group/unvaccinated group (l = 1 to 2), genotypem is the fixed-effect of genotype (m = 1 to 2), timen is the fixed-effect of time points of measurement prior and after vaccinations (m = 1 to 8), sexo is the fixed-effect of sex (o = 1 to 2), ANIMALijklmno is the random effect of animal, (genotype× time)mn is the interaction between genotype and time point, and ɛijklmno is the residual error.
RESULTS AND DISCUSSION
Single nucleotide polymorphisms
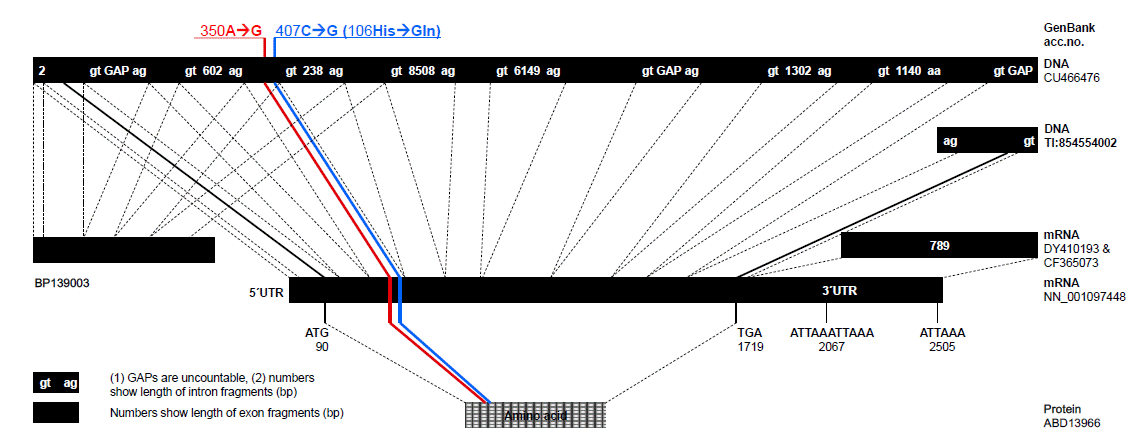
The whole cDNA length of the pC9 gene was sequenced (GenBank NM_001097448) containing a total of 2,536 bp encoding 543 amino acids (Figure 1). An alignment of cDNA sequence to NCBI Map Viewer revealed the pC9 had a long 3′untranslated region of 815 bp, which may play a role in C9 expression (DiScipio et al., 1984).
It was easy to identify three distinct structural units in the pC9 protein sequence, which have the characteristic features of the modules of thrombospondin type I (TSP1, amino acids 45–95), low-density lipoprotein receptor domain Class A (LDLa, amino acids 100–136), and MAC/perforin domain (MACPF, amino acids 297–507) according to SMART software (http://smart.embl-heidelberg.de/). The terminal complement components C6, C7, C8A, C8B, and C9 are structurally related proteins, differing in size and complexity. Actually, the pC9 protein shares 24, 22, 27, and 24% identity with pC6 (GenBank ABD13967), pC7 (GenBank AAD45918), pC8A (GenBank ABD13968), and pC8B (GenBank ABD13969), respectively. Probably, proteins of the terminal pathway (C6-9) originated from a single ancestral gene (Katagiri et al., 1999; Mondragón-Palomino et al., 1999). In various animal species, the LDLa domain contains the highly conserved sequence Asp-Cys-X-Asp-Gly-Ser-Asp-Glu, which is implied as a functional sequence for binding to apoprotein B and E (Yamamoto et al., 1984; Stanley et al., 1985).
Comparative sequencing of samples from different pig breeds revealed two SNPs at nucleotide 350A→G (codon 87CAA→CAG, TSP1 domain, exon 3) and 407C→G (codon 106CAC→CAG, LDLa domain, exon 3) (Figure 2). The first SNP is a silent mutation, whereas the second SNP has an amino acid exchange His→Gln. Genotyping was done in unrelated commercial animals of breeds LR, PIE, and MK as well as in the F2 DUMI population (Table 2).
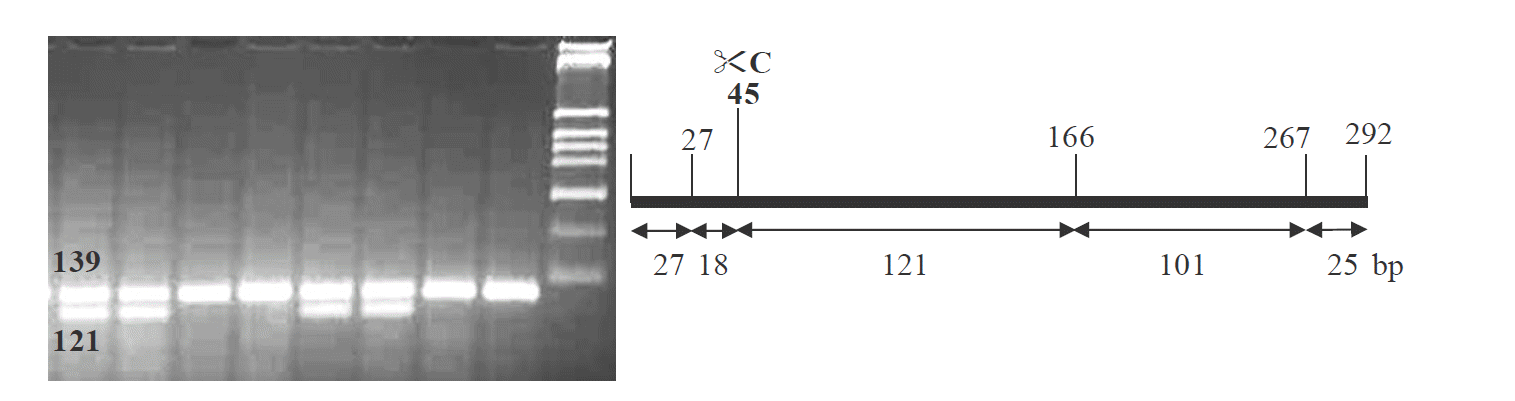
Representative pattern of detection of SNP using PCR-RFLP/HpyCH4III at position 407C→G, homologous GG (139 bp) and heterologous CG (121 bp+139 bp) in the pC9 candidate gene. SNP, single nucleotide polymorphism; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism.
At the 350A→G, LR, and PIE showed numerous AA homozygous and AG heterozygous genotypes whereas MK were fixed for GG homologous genotype. Another SNP site 407C→G was segregating in the F2 DUMI animals but not in the commercial animals (LR, PIE, or MK). No significant differences between the observed and the expected numbers of genotypes were found in LR, PIE, or MK populations for the pC9 gene. All genotypic and allelic patterns were in Hardy-Weinberg equilibrium.
In this study, the SNP with amino acid substitution was investigated for association with hemolytic complement activity in the classical and alternative pathway using the F2 DUMI resource population. This SNP showed two different genotypes GG and CG in F2 DUMI. The allele “C” maybe derived from Saddleback pigs, one of the breeds used to create Berlin Miniature pigs. These genetic variations may play an important role in the structural formation and function of the protein domains and therefore may affect the activity of the complement components in cell lysis.
Association
Significant difference between different genotypes GG (69.49±3.17 U/mL) and CG (63.59±3.65 U/mL) in hemolytic activity in the classical pathway (p = 0.049) was recorded. However, this was not found between two genotypes (respectively, 62.73±2.82 and 61.52±2.39 U/mL) in alternative pathway (Figure 3).
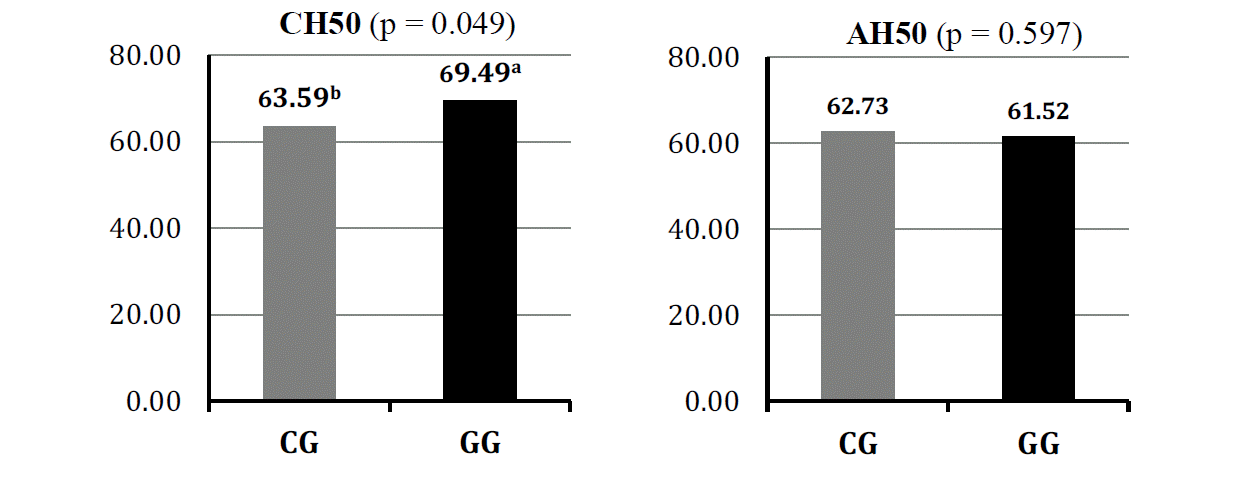
Plot of least square means of hemolytic complement activity in the classical (CH50) and alternative (AH50) pathway depending on genotypes of the porcine C9 gene.
Analyzing interaction between genotypes and hemolytic activity in the classical pathway indicated that the homozygous GG genotype always had higher hemolytic activity than the heterozygous CG during the experiment (Figure 4). Furthermore, CH50 reached a maximum level at day 4 after immunization with ADV for both homozygous GG (83.09±3.85 U/mL) and heterozygous CG (78.12±4.72 U/mL) genotypes. This can partly be due to higher age at ADV vaccination (14 weeks) compared to Mh vaccination (6 weeks of age) and the kind of vaccine. PRRS vaccination was only 2 weeks after ADV. Here a similar high complement activity was observed and an aging effect is not likely. However, this interaction in the classical pathway was not significant different (p = 0.314).

Plots of least squares means of hemolytic complement activity in the classical (CH50) and alternative (AH50) pathway along vaccination for the interaction of C9 genotypes and eight different time points (U/mL).
The results also demonstrated that there was significant difference in the interaction between genotypes and immunized point times in the alternative pathway (p = 0.034) (Figure 4). A linear increment of AH50 value along the experimental process was found.
Performance of hemolytic complement activity via the classical and alternative pathway can originate from many different causes including genetic variation within the pC9 candidate gene, in which the allele “G” at point mutation 407C→G was favourable. This SNP could be added to SNP-GeneChip for selecting healthy pigs, who may have naturally improved defence against invading pathogens.
Previous studies indicated that hemotylic complement activity was affected by many factors. For example, lipopolysaccharide of Mycoplasma species interacting with C1 and inducing an antibody-independent activation of the complement system via the classical pathway (Bredt et al., 1977; Rosendal et al., 1994), Epstein-Barr-Viruses activating both complement pathways (Mayes et al., 1984; Mold et al., 1988), vaccination affecting acute phase response through cytokines like interleukin-1 (IL1), IL6, and transforming growth factor beta 1 (TGFB1) that are released during immune response and, in particular, trigger the complement activity (Castell et al., 1989; Mackiewicz et al., 1990; Gonzalez-Ramon et al., 2000). Clearly, the activation of the complement system via the classical pathway is initiated by the interaction of the C1q component with the Fc regions of antigen-antibody complexes (Reid and Porter, 1981; Wimmers et al., 2003) while the alternative complement pathway is directly activated via binding of spontaneously activated C3b to the surface of a pathogen (Reid and Porter, 1981; Thacker, 2003). The complement system is immature at birth, then repeated antigenic stimulation leads to the complete maturation of specific immunity. Breed and age are important factors influencing the response to various stressors or infectious challenges (Sutherland et al., 2005). In this study, the hemolytic complement activity in the classical was stronger than in the alternative. It could be due to presence of the core motif Glu-X-Lys-X-Lys (E398IKGK and E442LKEK) in molecular structure of the pC9. Miletic and Frank (1995) suggested that C1q binding by immunoglobulin G utilized this motif. The last contribution may depend on both antigen-specific and antigen-nonspecific immune responses of host along the experiment and the antibodies that were accumulated after each vaccination causing responsiveness.
IMPLICATIONS
Genetic variation, kind of inoculated vaccine, aging, and the interaction among other complement components are major features causing either indirect or direct effects on complement activation of the both pathways. These make boosting hemolytic complement activity in the classical pathway significantly higher than in the alternative pathway. Complement activation via viral/bacterial vaccination is very complicated. The obtained results provide the means for further understanding the role of the pC9 in natural immune response of the host against pathogens. It also promotes the pC9 as candidate gene for disease resistance.
ACKNOWLEDGMENTS
We greatly appreciate support from the BMBF (Bundesministerium für Bildung und Forschung) and DRL (Deutschen Zentrums für Luft- und Raumfahrt) to conduct this project.