Comparison of Faecal Microbial Community of Lantang, Bama, Erhualian, Meishan, Xiaomeishan, Duroc, Landrace, and Yorkshire Sows
Article information
Abstract
The objective of this study was to investigate differences in the faecal microbial composition among Lantang, Bama, Erhualian, Meishan, Xiaomeishan, Duroc, Landrace, and Yorkshire sows and to explore the possible link of the pig breed with the gut microbial community. Among the sows, the Meishan, Landrace, Duroc, and Yorkshire sows were from the same breeding farm with the same feed. Fresh faeces were collected from three sows of each purebred breed for microbiota analysis and volatile fatty acid (VFA) determination. Denaturing gradient gel electrophoresis (DGGE) analysis revealed that samples from Bama, Erhualian, and Xiaomeishan sows, which from different farms, were generally grouped in one cluster, with similarity higher than 67.2%, and those from Duroc, Landrace, and Yorkshire sows were grouped in another cluster. Principal component analysis of the DGGE profile showed that samples from the foreign breeds and the samples from the Chinese indigenous breeds were scattered in two different groups, irrespective of the farm origin. Faecal VFA concentrations were significantly affected by the pig breed. The proportion of acetate was higher in the Bama sows than in the other breeds. The real-time PCR analysis showed that 16S rRNA gene copies of total bacteria, Firmicutes and Bacteroidetes were significantly higher in the Bama sows compared to Xiaomeishan and Duroc sows. Both Meishan and Erhualian sows had higher numbers of total bacteria, Firmicutes, Bacteroidetes and sulphate-reducing bacteria as compared to Duroc sows. The results suggest that the pig breed affects the composition of gut microbiota. The microbial composition is different with different breeds, especially between overseas breeds (lean type) and Chinese breeds (relatively obese type).
INTRODUCTION
The assortment of microorganisms that inhabit the mammalian gastrointestinal tract is known as the gut microbiota, and the composition of this microbial community is host specific, evolving throughout an individual’s lifetime. The gut microbiota can be considered a microbial organ placed within a host organ, which has a central position in health and disease (Bäckhed et al., 2005). This ‘organ’ is intimately involved in numerous aspects of normal host physiology, from nutritional status to behaviour and stress responses. It consumes, stores and redistributes energy, it mediates physiologically important chemical transformations, and it can maintain and repair itself through self-replication (Bäckhed et al., 2005; Sekirov et al., 2010). The community is dominated by anaerobic bacteria, which process plant polysaccharides and indigestible components of the host diet, such as cellulose, hemicellulose and resistant starch, into simple sugars, short chain fatty acids (SCFAs) and other nutrients beneficial to the host. Microorganisms obtain rich polysaccharides as their survival substrate in the anaerobic environment provided by the host, and the host has access to a source of carbon and energy from symbiotic activities (Bäckhed et al., 2004; Bäckhed et al., 2005).
China has the largest swine industry in the world. Due to variations in the natural environment and differences in socio-economic conditions, more than 100 local swine breeds are reared in China (Li et al., 2011). The Chinese native pig breeds exhibit many similar properties. For example, they all have high reproductive rates and are adaptable to extensive feeding. They also have an excellent ability to utilize roughage and have good meat quality. Although China has hundreds of pig breeds, only a few breeds of hogs are widely used in commercial pork production, along with some common foreign breeds, including Yorkshire, Duroc, Landrace, and Pietrain. This is because of their undesirable traits, such as slow growth rate, low dress percentage and low lean meat percentage. In recent years, the importance of the gut microbiota to the body’s energy metabolism has been widely acknowledged (DiBaise et al., 2008; Schwiertz et al., 2010). Obesity has recently been linked to the composition of microbiota and the production of SCFAs, and recent reports of a possible correlation between the gut microbiota and obesity have placed the focus on the significance of the microbiota in wellbeing (Bäckhed et al., 2004; Turroni et al., 2008). Genome research has shown that the gut microbiota is enriched in the metabolic pathways associated with glucose metabolism (Nicholson et al., 2012). Gut microorganisms are mainly composed of Bacteroidetes and Firmicutes divisions (DiBaise et al., 2008). They comprise more than 90% of all phylogenetic types in the pig gut (Leser et al., 2002; Guo et al., 2008). In addition, methanogens have also been reported as dominant in pigs (Mao et al., 2011; Luo et al., 2012). Firmicutes, Bacteroidetes and methanogens are thought to play a role in the energy metabolism of the host (Schwiertz et al., 2010). Methanogens also play an important role in maintaining normal fermentation in the pig hindgut (Samuel and Gordon, 2006).
Pigs have a strong capacity for fat storage. The fat storage capacity of Chinese native species such as Meishan and Erhualian, which are genetically obese, is different to that of foreign breeds. In contrast, Duroc, Landrace and Yorkshire breeds are typically lean breeds. Variations in the intestinal microbial composition of different breeds of sows have rarely been reported. Therefore, the current study compared the faecal microbial community between different pig breeds including Lantang, Bama, Erhualian, Meishan, Xiaomeishan, Duroc, Landrace, and Yorkshire sows, with special focus on the comparisons between Chinese breeds and overseas breeds. The faecal microbial community of the eight pig breeds was compared by denaturing gradient gel electrophoresis (DGGE) and real-time PCR analysis.
MATERIALS AND METHODS
Animals and sampling
Three healthy adult purebred sows from each of the Lantang, Bama, Erhualian, Meishan, Xiaomeishan, Duroc, Landrace, and Yorkshire breeds were sampled for collection of faeces. The pigs were from different regions in China and raised on separate farms. Based on previous studies (Mo et al., 2003; Guo et al., 2008; Schwiertz et al., 2010), we collected some sow samples from different farms. In order to make comparisons of different breeds under the same conditions, we also selected a farm where we were able to collect four different breed sow samples. All sows had littered 2–3 times and were not in pregnancy during the sampling time. All sows were fed with diets based on corn and soybean meal. The Bama and Lantang sows were from Guangxi and Guangdong provinces respectively, and others from Jiangsu province. All samples were collected randomly from breeding farms, with conditions listed in Table 1.

Chinese indigenous pig breeds and introduced pig breeds included in the study and their sampling location
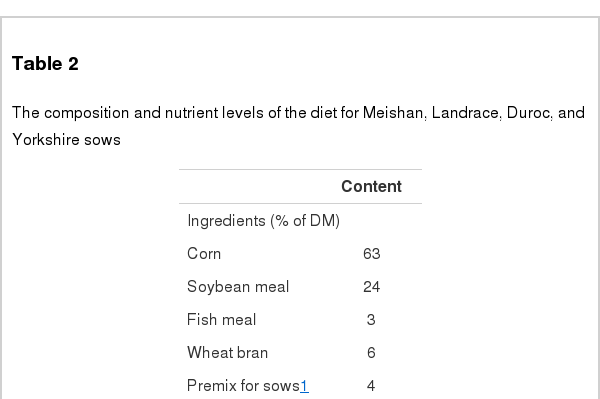
Meanwhile, the Meishan, Landrace, Duroc, and Yorkshire sows were housed in the same room on a breeding farm in Taicang City and fed with diets based on corn and soybean meal as well (Table 2). The breed was the only variance among them, which was to ensure the differences in the faecal microbial composition was due to pig breed. All pigs had ad libitum access to diet and water. All samples were collected within a month in March 2012. On the day of sampling, fresh faecal samples (approximate 150 g) were immediately collected before feeding in the morning and then immediately transported to the laboratory in foamed plastic containers with dry ice, and stored at −20°C until analysis.
Volatile fatty acid analysis
Faecal samples were prepared for volatile fatty acid (VFA) analysis by mixing 0.4 g of faeces with 0.2 mL of 25% (w/v) metaphosphoric acid and 2 mL of water. The samples were then centrifuged (17,000×g for 10 min), and supernatant fluid was used for VFA determination (Shimadzu, GC-14A, Japan) (Mao et al., 2013).
DNA extraction and PCR amplification
Total bacterial DNA was extracted from each faecal sample (0.3 g) using the bead-beating method with a mini-bead beater (Biospec Products, Bartlesville, OK, USA), followed by phenol-chloroform extraction (Zoetendal et al., 1998). The DNA was then precipitated with ethanol, and the pellets were dissolved in 50 μL of Tris EDTA (TE). Primers U968-GC and L1401 (Nubel et al., 1996) (Table 3) were used to amplify the V6 to V8 variable regions of the bacterial 16S rRNA gene. PCR was performed with the Taq DNA polymerase kit from Promega (Madison, WI, USA). PCR amplification was performed using the following program: 94°C for 7 min and 35 cycles of 94°C for 30 s, 56°C for 20 s, 68°C for 40 s and 68°C for a 7 min final extension. Aliquots of 5 μL PCR products were analysed by electrophoresis on 1.2% agarose gel (w/v) containing GoldView to check the size and the amount of the amplicons.
DGGE analysis
PCR amplicons obtained from the V6 to V8 regions of the 16S rRNA genes were separated by DGGE according to the specifications of Muyzer et al. (1993) using a Dcode TM system (Bio-Rad, Hercules, CA, USA). DGGE was performed in 8% polyacrylamide gels containing 37.5:1 acrylamide-bisacrylamide and a denaturing gradient of 38% to 53%. Electrophoresis was initiated by prerunning for 10 min at 200 V and subsequently continued at a fixed voltage of 85 V for 12 h at 60°C. The gel was stained with 0.2% AgNO3 after completion of electrophoresis. The DGGE gels were scanned using GS-800 Calibrated Densitometer (Bio-Rad) and Molecular Analyst version 1.61 software (Bio-Rad). Similarity analysis, based on the unweighted pair group mean average (UPGMA), and principle component analysis (PCA) were performed using GelCompar II (Applied Maths, Gent, Belgium) and Canoco software (Microcomputer Power, Ithaca, NY, USA). The Shannon index of general diversity, H′, was calculated to determine the structural diversity of the bacterial community. H′ was calculated using the following function: H′ = −∑Pi ln Pi, where Pi is the importance probability of the bands in a lane. The importance probability, Pi, was calculated as Pi = ni/H′, where ni is the height of a peak and H′ is the sum of all peak heights in the densitometric curve.
Real-time PCR assay for quantification of total bacteria, Bacteroidetes, Firmicutes, methanogens, and sulphate-reducing bacteria
Real-time PCR was performed on a StepOnePlus (Applied Biosystems, California, USA) with StepOne Software (version 2.2.2, Applied Biosystems). The real-time PCR reaction mixture (20 μL) consisted of 10 μL of IQ SYBR Green Supermix (Bio-Rad), 0.4 μL of each primer set, 0.4 μL ROX Reference Dye, 2 μL of template DNA and 6.8 μL PCR-grade H2O. Quantitative PCR amplification was performed using the following conditions: 95°C for 30 s and 40 cycles of 95°C for 5 s, 60°C for 30 s and one cycle of 95°C for 15 s, 60°C for 1 min and 95°C for 15 s. Quantification of 16S rRNA gene copies in each sample was performed in triplicate, and the mean value was calculated. Standard curves were generated with 10-fold serial dilutions of the 16S rRNA genes amplified from the respective target strains. The concentration of 16S rRNA gene abundance was plotted against the CT value. All the PCR primers used in this study are listed in Table 3. Universal primers, Bact1369F and Bact1492R (Suzuki et al., 2000), were used to estimate the total number of copies of the bacterial 16S rRNA gene in each sample. According to the method of Guo et al. (2008), the 16S rRNA genes of Bacteroidetes and Firmicutes were quantified by primers Bact934F/Bact1060R and Firm934F/Firm1060R, respectively. Methanogens and sulphate-reducing bacteria (SRB) were quantified using the primers Methanogen-F/Methanogen-R (Denman et al., 2007) and DsrA-F/DsrA-R (Devkota et al., 2012) respectively.
Statistical analysis
All the statistical analysis was performed using SPSS 17.0 software (SPSS Inc., Chicago, IL). Statistically significant differences were determined by the one-way ANOVA test. To assess the mean differences in the breeds, a univariate analysis of variance was used, followed by Tukey’s post hoc multiple comparison test. p-values <0.05 were considered significant.
RESULTS
Concentrations of volatile fatty acids in different pig breeds
The differences in the stool VFA concentrations among the eight breeds from different areas were considerable (p< 0.05) (Table 4). Acetate, propionate, total VFA (TVFA) and branched chain fatty acid (BCFA) concentrations were significantly higher in the Erhualian breed than in the Xiaomeishan, Duroc, and Meishan breeds. The concentration of butyrate was significantly higher in the Erhualian and Landrace breeds than in the Duroc and Meishan breeds. Among Meishan, Landrace, Duroc and Yorkshire breeds, which were raised on a commercial farm in Taicang City using the same feed and under the same environmental conditions, the highest concentration of acetate, propionate, butyrate and TVFA was observed in the Landrace sows (Table 4), with their values greater than those in the Duroc and Meishan sows. The concentration of BCFA was higher in the Landrace sows than in the Meishan sows.

Concentrations of VFA (m mol/kg wet wt) in the faecal samples of the different pig breeds from different areas
As shown in Table 5, the proportion of acetate was higher in the Bama sows than in the other sows. The proportion of propionate in the Erhualian sows was significantly higher than in the Lantang, Bama, Xiaomeishan and Yorkshire sows. Comparison of the proportions of butyrate revealed a considerable decrease in the Bama sows compared to the levels in the Meishan, Erhualian, Duroc, and Yorkshire sows. The ratio of acetate and propionate was higher in the Bama sows than in the Erhualian, Meishan, Duroc, and Landrace sows. In addition, compared to the Lantang, Xiaomeishan and Yorkshire sows, the Erhualian sows had a lower ratio of acetate and propionate.
DGGE analysis of bacteria diversity in faecal samples
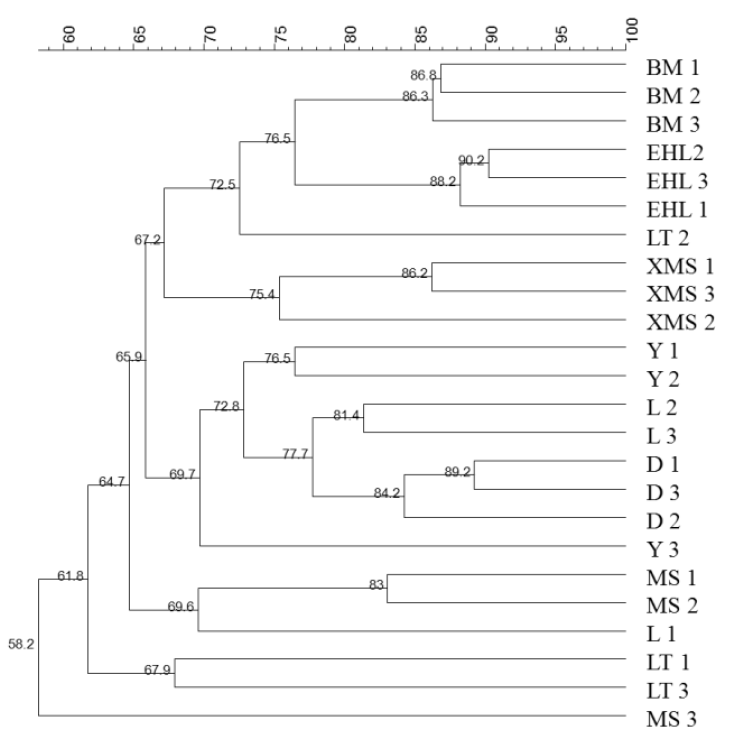
The DGGE profiles of the PCR products of the V6-V8 regions of the 16S rRNA genes from the faecal samples revealed remarkable differences among the eight pig breeds. As shown in Figure 1, the samples from the Landrace, Yorkshire, and Duroc sows had similar band pattern with high similarity, while the samples from the Lantang, Bama, Erhualian, Xiaomeishan, and Meishan sows showed similar band pattern, but with relatively low similarity. Among the Chinese breeds, Bama sows showed some unique bands that were absent in other breeds.
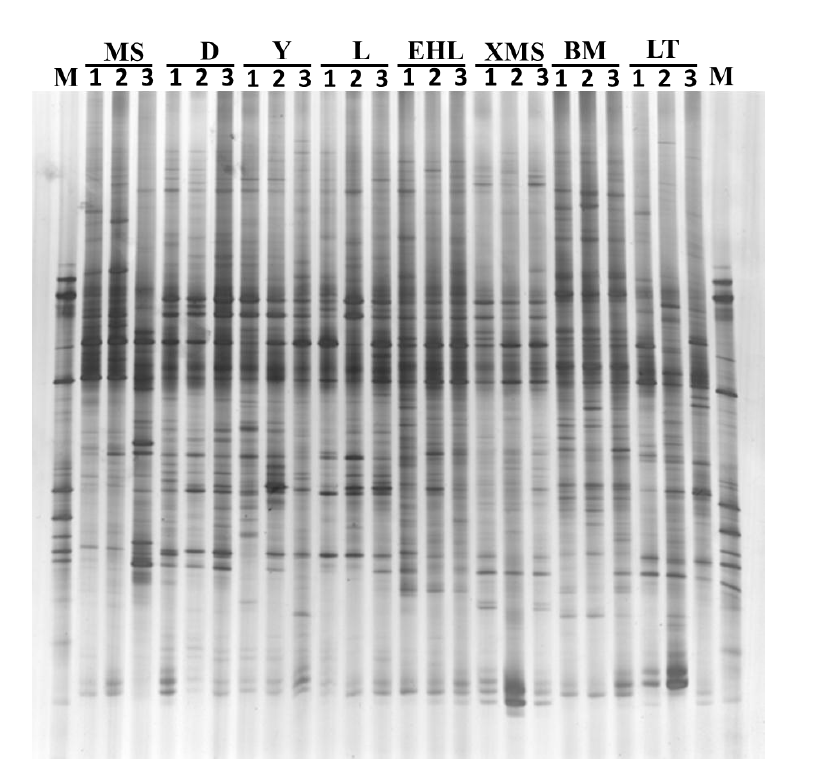
DGGE profiles of the V6–V8 regions of the 16S rRNA gene from faecal bacteria of different pig breeds. M, Marker; MS 1–3, Meishan; D 1–3, Duroc; Y 1–3, Yorkshire; L 1–3, Landrace; LT 1–3, Lantang; BM 1–3, Bama; EHL 1–3, Erhualian; XMS 1–3, Xiaomeishan.
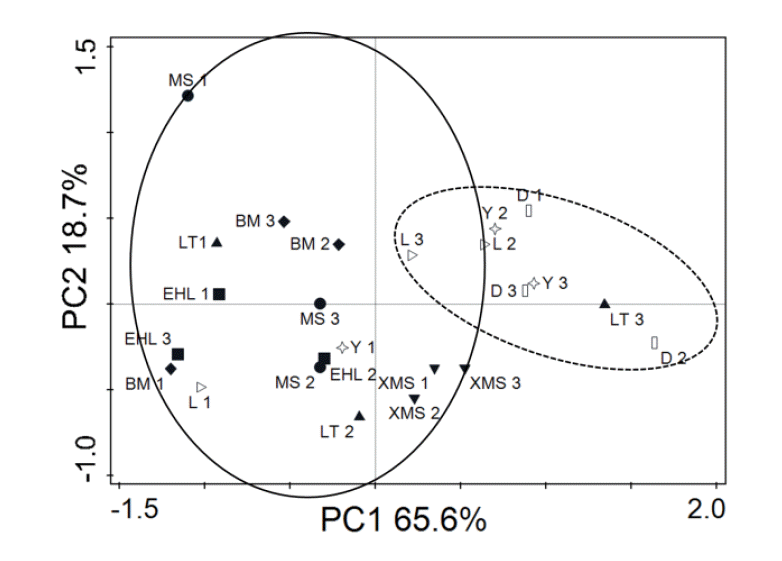
To obtain an objective interpretation of the electrophoretic patterns of the groups, the samples were subjected to a numerical analysis based on the Dice similarity coefficient, followed by cluster analysis (Figure 2). Samples from the Bama, Erhualian and Xiaomeishan sows grouped in a cluster with similarity indices higher than 67%, and samples from the Duroc, Landrace, and Yorkshire sows were grouped in another cluster with similarity indices higher than 69%. The PCA analysis of the DGGE profile further showed that the first principal component (PC1), which explained 20.2% of the variation, separated the bacterial communities in the faeces of the Duroc, Landrace, and Yorkshire from local pig breeds (Figure 3).
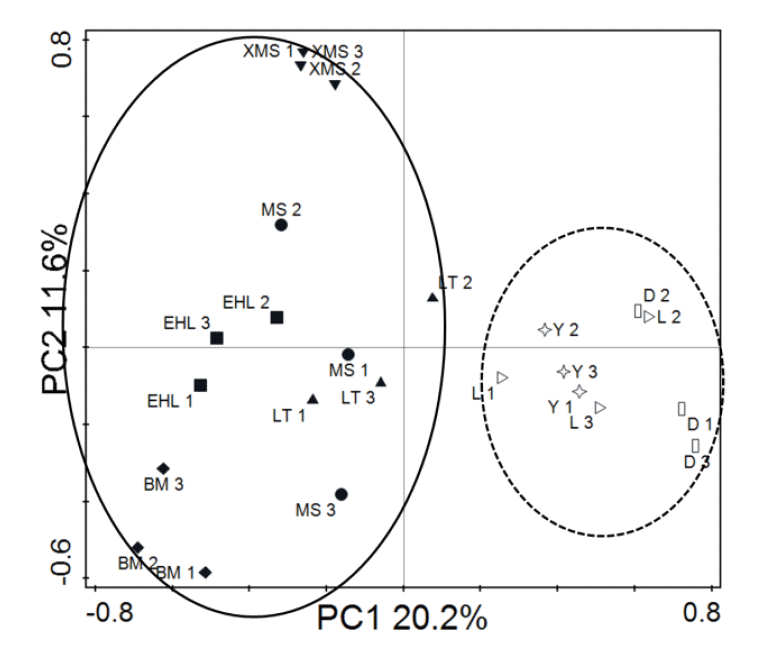
PCA analysis of DGGE profiles from faecal bacteria of the different pig breeds. PC1 illustrates the difference in sample L 1–3, Y 1–3, and D 1–3 from the rest of the samples (20.2% of the variation), and PC2 illustrates the difference in sample XMS 1–3 (11.6% of the variation).
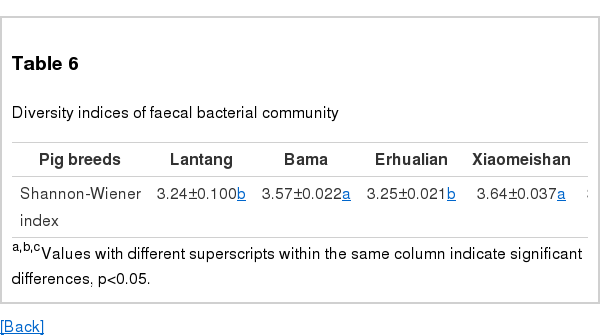
Shannon diversity indices were calculated to compare the diversity of the dominant bacterial microbiota in the faeces of the different pig breeds (Table 6). Statistical analysis showed that the index was significantly different between the Bama and Xiaomeishan sows and between the Lantang, Erhualian, Meishan, Duroc, Landrace, and Yorkshire sows.
Quantification of the predominant bacterial groups in the stool samples
As shown in Table 7, the 16S rRNA gene copies of the total bacteria in the Bama pigs were significantly higher than those in the Erhualian, Landrace, Yorkshire, and Duroc sows. In addition, the total bacteria concentrations in the Erhualian and Duroc sows were significant higher than in the Meishan and Xiaomeishan sows. The Bama and Meishan pigs harboured significantly higher faecal concentrations of Firmicutes than the Erhualian and Duroc sows. Both the Meishan and Erhualian sows exhibited higher faecal concentrations of Firmicutes compared to the Duroc sows. Copies of Bacteroidetes in the Bama, Meishan and Xiaomeishan sows were significantly higher than those in the Erhualian, Yorkshire, and Duroc sows. On the other hand, copies of methanogens in the Duroc sows were significantly lower than in the other sows. In addition, Xiaomeishan pigs harboured significantly higher faecal concentrations of SRB than the Yorkshire and Duroc sows, and the Duroc sows harboured significantly higher faecal concentrations of SRB than the Lantang, Bama, and Meishan sows.
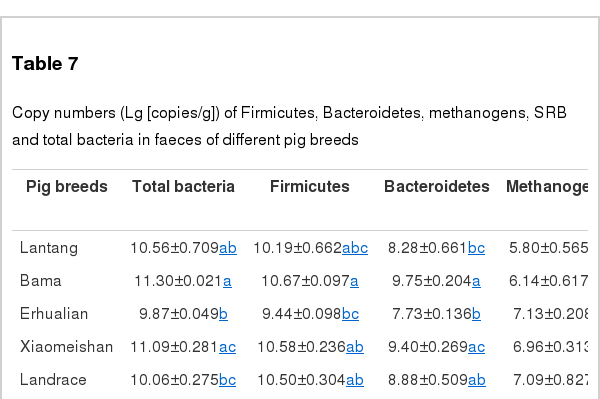
Copy numbers (Lg [copies/g]) of Firmicutes, Bacteroidetes, methanogens, SRB and total bacteria in faeces of different pig breeds
The PCA analysis of the numbers of the predominant bacteria and methanogens showed that samples from the Bama, Erhualian, Xiaomeishan, and Meishan sows clustered tightly (Figure 4). The samples from the Duroc, Landrace and Yorkshire sows were separated from the others by the first principal component (PC1) and the second principal component (PC2). The results indicate that samples from foreign breeds and samples from indigenous pig breeds are mainly scattered into two different groups.
DISCUSSION
The results of VFA, DGGE, and real-time PCR showed great differences in both the bacterial communities and VFA profiles between the Bama pigs and the others, suggesting relations between VFA and microbiota (Fernandes et al., 2013; Timm et al., 2013). Bama pigs are a miniature breed, and their phenotype differs greatly compared to that of the other breeds. These results suggest that gut microbes may be related to the phenotype of the breed.
Previous research showed that intestinal microbiota can be affected by many factors, including genetics, dietary composition, age, living environment and stress (Mueller et al., 2006; Spor et al., 2011; Xu et al., 2013). In this study, most of the selected breeds were from different regions, where the living environment is different. These factors, in addition to the aforementioned variables, may explain the differences in the microbiota. With respect to their living environment, the geographical environment of living conditions of the Bama and Lantang sows, which were from Guangxi and Guangdong provinces, were more similar to each other, while very different from other sows. In contrast to our expectations, the microbiota of the Bama and Lantang was significantly different. Likewise, the Xiaomeishan, Erhualian, Meishan, Landrace, Yorkshire, and Duroc sows shared a similar geographical setting, but the microbiota in them are differed, although four of the breeds (Meishan, Landrace, Yorkshire, and Duroc) came from the same farm, were fed the same diet and shared in the same rearing environment.
The PCA analysis indicated that samples from foreign breeds and samples from Chinese breeds are mainly scattered into two different regions. The Landrace, Yorkshire and Duroc sows are introduced species, although of different origin, they have some similarly desirable traits, such as a high growth rate, high dress percentage and high lean meat percentage. The dominant microbial community of these overseas breeds shared higher similarities while the Chinese breeds had relatively low similarity. The results indicated that the microbial community of these fast growing overseas pigs share very common microbiota.
Methanogens play an important role in maintaining normal fermentation in the pig hindgut (Samuel and Gordon, 2006). As the major energy producers of the gut, Bacteroidetes and Firmicutes break down various types of otherwise indigestible polysaccharides, including plant-derived pectin, cellulose, hemicellulose and resistant starches. They ferment the resulting monosaccharide into SCFAs, principally acetate, propionate and butyrate, as well as other organic acids such as hydrogen (H2) and carbon dioxide (CO2). Accumulation of H2 inhibits bacterial NADH dehydrogenases, thereby reducing the yield of ATP. Methanogens can remove H2 and CO2 and synthesise methane to improve the fermentation efficiency (Samuel and Gordon, 2006). Therefore, the kind and quantity of VFA can significantly affect methanogens. SRB are a complex bacterial group that contribute to a variety of essential functions in many anaerobic environments. The toxic gas, hydrogen sulphide (H2S) is generated by SRB and linked to chronic intestinal disorders. H2S selectively impairs the oxidation of n-butyrate by colonic epithelial cells. Many intracellular processes depend on the oxidation of n-butyrate in membrane lipid biosynthesis, ion absorption, mucin synthesis and detoxification processes in colonocytes, and diminished n-butyrate metabolism is likely to compromise the epithelial cell barrier (Deplancke et al., 2000). The genetic background could have a substantial effect on fat deposition. Chinese native pigs have a strong capacity for fat storage compared with foreign pigs. Recent studies showed that body fat storage affects the gut microbial ecology (Ley et al., 2005; Ley et al., 2006; Turnbaugh et al., 2006). However, the mechanism of interactions between body fat storage and the gut microbial community is not yet clear. Based on the results of DGGE and real-time PCR in the current study, it seems that the pig breed could influence the faecal microbial composition.
In this study, except for the Xiaomeishan sows, the 16S rRNA gene copies of the total bacteria, Firmicutes, Bacteroidetes, and SRB bacteria in the Chinese native pig breeds were higher than those in the foreign breeds. Meishan, Lantang, Bama, and Erhualian sows have abundant microbiota, which are involved in the digestion of food and the stimulation of the development of the immune system to prevent damage to the body. In the present research, all the sows had a low percentage of Firmicutes (based on all bacteria) (data not shown). Leser et al. (2002) reported the percentage of Firmicutes were 81%, of all phylogenetic types in pigs and Guo et al. (2008) found the percentage of Firmicutes in Meishan pigs (72.09%) was higher in numerical value than in Landrace pigs (61.17%). In our study, the percentages of Firmicutes in Chinese domestic breeds were generally lower than in the foreign pig breeds, even in the same farm, where the percentage of Firmicutes in the Meishan sows (64.44±2.9%) was lower than in the Landrace (73.02±9.8%) and Yorkshire sows (78.46±10.9%), but higher than in the Duroc sows (57.08±7.1%). Although the Duroc, Landrace, and Yorkshire breeds are all lean type of pigs, there were differences among their microbiota, with the number of copies of Firmicutes and methanogens in the Landrace sows being significantly higher than in the Duroc sows. Recent studies have confirmed a relationship between fat deposition and the gut microbial community of Firmicutes, Bacteroidetes, and methanogens (Turnbaugh et al., 2006; Tremaroli and Bäckhed 2012). However, the specific relationship is still poorly understood, and mechanisms between microbiota and fat deposition should be further studied.
CONCLUSION
The results suggest that the composition of the pig gut microbiota is different with different breeds, especially between overseas breeds (lean type) and Chinese breeds (relatively obese type). Within the Chinese breeds, the gut microbial community of the Bama breed, the miniature breed, was apparently different from other pig breeds. These differences may be associated with their conformation and growth characteristics. However, the underlying relationship between gut microbiota and growth performance of the pig needs further extensive studies.
ACKNOWLEDGEMENTS
This work was supported by the Natural Science Foundation of China (NSFC) (30810103909) and China-EU cooperation project (1008), and the European Union through the European Frame Work 7 project ‘INTERPLAY’ (Project No. 227549).