Tissues Expression, Polymorphisms Identification of FcRn Gene and Its Relationship with Serum Classical Swine Fever Virus Antibody Level in Pigs
Article information
Abstract
Neonatal Fc receptor (FcRn) gene encodes a receptor that binds the Fc region of monomeric immunoglobulin G (IgG) and is responsible for IgG transport and stabilization. In this report, the 8,900 bp porcine FcRn genomic DNA structure was identified and putative FcRn protein included 356 amino acids. Alignment and phylogenetic analysis of the porcine FcRn amino acid sequences with their homologies of other species showed high identity. Tissues expression of FcRn mRNA was detected by real time quantitative polymerase chain reaction (Q-PCR), the results revealed FcRn expressed widely in ten analyzed tissues. One single nucleotide polymorphism (SNP) (HQ026019:g.8526 C>T) in exon6 region of porcine FcRn gene was demonstrated by DNA sequencing analysis. A further analysis of SNP genotypes associated with serum Classical Swine Fever Virus antibody (anti-CSFV) concentration was performed in three pig populations including Large White, Landrace and Songliao Black pig (a Chinese indigenous breed). Our results of statistical analysis showed that the SNP had a highly significant association with the level of anti-CSFV antibody (At d 20; At d 35) in serum (p = 0.008; p = 0.0001). Investigation of expression and polymorphisms of the porcine FcRn gene will help us in further understanding the molecular basis of the antibody regulation pathway in the porcine immune response. All these results indicate that FcRn gene might be regarded as a molecular marker for genetic selection of anti-CSFV antibody level in pig disease resistance breeding programmes.
INTRODUCTION
Immunoglobulin G (IgG) is the most prevalent antibody that has an important role in long-term humoral immunity. It has also been regarded as a pathological effecter of systemic autoimmune diseases such as systemic lupus erythematosus and spontaneous abortion in multiparous females (Roopenian et al., 2003). It has long been known that IgG is the only class of antibodies that is efficiently transferred from mother to offspring to develop short-term passive immunity in new born animals (Morphis and Gitlin, 1970; Brambell, 1996; Roopenian and Akilesh, 2007). This specific transport of IgG is dependent on the neonatal Fc receptor (FcRn) (Simister and Mostov, 1989; Roberts et al., 1990), which was originally identified by Simister and Waldmann (Jones and Waldmann, 1972).
FcRn gene functions as the receptor responsible for IgG binding and shares 22 to 29% sequence identity with Major Histocompatibility Complex (MHC) Class I molecules (Simister and Mostov, 1989; Raghavan and Bjorkman, 1996). In this process, FcRn binds to the Fc region of IgG in a strictly pH-dependent manner (Brambell, 1966; Praetor et al., 1999) and is then transported through maternally derived IgG from milk across the intestinal epithelial-cell layer of the neonatal rodents. In addition, beyond the perinatal period, FcRn protects IgG from degradation, thereby prolongs the half-life of IgG antibodies in the serum, this function is key to IgG homeostasis, that helps this protective class of antibody in maintaining a high concentration in immune circulation (Jones and Waldmann, 1972; McCarthy et al., 2000; Roopenian et al., 2003; Roopenian and Akilesh, 2007).
Since FcRn is the only receptor responsible for perinatal IgG transport, FcRn has been regarded as a promising target gene for enhancing protective humoral immunity. Due to the important role of FcRn for IgG transport and stabilization, genetic variations of FcRn-dependent transport may influence antibody immunity of the newborn. Strong evidence reveals various mutations at different positions of some amino acid residues around the FcRn-Fc binding interface. These improve the pH-dependent binding of human IgG to FcRn (Shields et al., 2001; Hinton et al., 2004; Kamei et al., 2005; Vaccaro et al., 2005; Hinton et al., 2006; Petkova et al., 2006). Therefore, research into the genetic mechanism of FcRn gene seems to be an attractive strategy and may provide reference for modulating specific IgG transport to promote host’s disease defense.
Considering its influence on the antibody immune response pathway, FcRn could be an important candidate gene for porcine antibody immune response and disease susceptibility. In this study, we firstly identified the genomic structure of the porcine FcRn gene, detected its expression in different tissues and polymorphisms in the regions of exons, and we also further performed an association analysis between the SNPs of the FcRn gene and anti-CSFV antibody level in serum to estimate their possible effects in three pig populations.
MATERIALS AND METHODS
Animals and DNA isolation
Three pig breed populations including Landrace, Large White and a Chinese indigenous breed Songliao Black were collected from the experimental farm of the Institute of Animal Sciences, Chinese Academy of Agricultural Sciences, Beijing, China. 296 pigs were vaccinated with CSF live vaccine at the age of 21 days. The first blood samples were collected from each piglet one day before the vaccination (At d 20), and 2 wks after the vaccination, the second blood samples were collected (At d 35). Ear tissues and blood samples of all pigs were also collected for DNA extraction using phenol/chloroform extraction and ethanol precipitation (Sambrook et al., 1989). Nine different tissues, including heart, liver, spleen, lung, kidney, skeletal muscle, lymph node, brain and thymus, were collected from three 35-d-old Landrace pigs, then immediately frozen in liquid nitrogen and stored at −80°C.
Measurement of CSFV antibody IgG level in serum
Serum samples from three pig populations were stored at −80°C. Anti-CSFV IgG level in serum samples were measured using the Herdchek* CSFV Antibody (IgG) ELISA Test Kit (IDEXX Laboratories, America), which contains 96-well test plates coated with CSFV antigen, sample diluent, positive control: standards of known CSFV antibody concentrations, negative control, washing solution, a CSFV antigen and a biotinylated monoclonal antibody specific to CSFV, and a tetramethyl benzidine (TMB) substrate solution. Serum samples from the pig populations and standards of known CSFV concentrations were loaded into the wells on the test plate: 50 μl of each serum sample or standard per well. The CSFV antigen and the biotinylated monoclonal antibody specific to CSFV were added to each well and were incubated at 20°C for 120 min. The wells were washed and with then 100 μl anti-CSFV horseradish peroxidase (HRPO) conjugate was added. After incubation at room temperature for 30 min, the wells were washed to remove unbound enzymes and the substrate solution, which reacted with the bound enzyme to induce a colour, was added. The intensity of the colour was proportional to the concentration of CSFV IgG present in the serum samples. The intensity of the colour was measured with an ELISA reader at 450 nm to calculate the mean absorbance values, these values were then converted into CSFV antibody concentration blocking percentage (%) in serum according to the manufacturer’s instructions.
Total RNA extraction and cDNA synthesis
The total RNA was extracted from ten different tissues (liver, thymus, spleen, lymphoid node, heart, skeletal muscle, kidney, lung, small intestine, and brain) of three 35 days old Landrace pigs with a TRIzol reagent (Invitrogen, USA). In order to prevent serious contamination by genomic DNA, DNaseI (Beijing Tiangen Biotechnology, China) treatment on the total RNA was carried out, and then the first strand cDNA was synthesized in the presence of 2 μg total RNA, 0.5 μM oligo(dT)18, 200 μM dNTPs, 10 U RNAsin (Promega, Madison, WI, USA), 1×M-MLV RT buffer, and 300 U M-MLV reverse transcriptase (Promega, USA) in a volume of 50 μl at 40°C for 1 h.
Real time Q-PCR analysis of FcRn mRNA expression
The real time quantitative PCR (Q-PCR) method was used to investigate the expression of FcRn gene in ten porcine tissues. A quantitative primer pair which spanned an intron was designed to eliminate potential confounding results from genomic DNA contamination (Table 1). Another primer set was also designed to amplify porcine glyceraldehyde3-phosphate dehydrogenase (GAPDH) mRNA (GenBank accession number: AF017079) as a positive control (F1 5′-TGAGACACGATGGTGAAGGT-3′ and R1 5′-GGCATTGCTGATGATCTTGA-3′). PCR reactions were performed in a final volume of 15 μl using DyNAmo™ SYBR® Green qPCR Kits (Finnzymes, Finland). All reactions were performed in optical tubes (Bio-Rad Laboratories, USA). First, 1 μl of template and 1 μl specific primer were delivered into the tubes, followed by 8.0 μl of SYBR® Green Q-PCR master mix and 6.0 μl ddH2O. The tubes were sealed with optical caps (Bio-Rad Laboratories, USA). All reactions were performed with a DNA Engine Opticon 2 Real-Time system (MJ Research, USA). The PCR program was optimized and consisted of an enzyme activation step (95°C, 10 min), forty cycles of denaturation (95°C for 30 s), annealing extension (60.5°C, for 30 s), extension (72°C for 35 s), reading plate, and incubation (84°C for 1 s), followed by an extension at 72°C for 10 min. Then, a melting curve from 55°C to 95°C, reading every 0.3°C for 1 s, was drawn. All data were analyzed using Opticon Monitor 2.0 software (MJ Research, USA).
Genomic organization and SNPs identification of the porcine FcRn gene
To characterize the genomic structure of the porcine FcRn gene, the mRNA sequences of the porcine FcRn gene (GenBank accession number: AY135635) was used to search for porcine homologous high throughput genomic sequences (HTGS) in the porcine genomic databases by BLAST (http://blast.ncbi.nlm.nih.gov) of NCBI GenBank database (http://www.ncbi.nlm.nih.gov/). One genomic DNA fragment (GenBank accession number: FP102630.3) was obtained, which contains all introns and exons of the porcine FcRn gene. Furthermore, the genomic organization was also subsequently deduced by experimental confirmation using exon-exon PCR across predicted introns or exon-intron PCR. Finally, the sequence of porcine FcRn genomic DNA was submitted to NCBI GenBank database.
The porcine FcRn gene consists of six exons. Therefore, using porcine genomic DNA, six primer pairs were designed for PCR amplification of DNA sequences including all exon regions of the IRF6 gene (Table 1). The DNA from two pigs from each breed was selected for PCR amplification. All PCR fragments were purified with a Gel Extraction Mini Kit (Beijing Tiangen Biotechnology, China) and then sequenced. After comparing the sequences obtained from the same PCR fragments from two pigs of each of the Landrace, Large White and Songliao Black breeds the polymorphism of the FcRn gene was found using DNAMAN software (version 5.2.10, Lyn-non Biosoft, Canada). Then all 296 samples from the three pig populations were genotyped by directly sequencing of PCR products.
Association analysis
In the end, we performed an association analysis of between genotypes of the SNP and serum CSFV antibody level by Statistical Analysis System (SAS) software (version 9.13), based on the following mixed model:
Where y is the vector of CSFV antibody IgG level in serum analyzed, X is the design matrix for fixed effects; β is the vector of fixed effects parameter including breed, ELISA plate effect and genotype effect. Z is the design matrix of random animal effects; b is the mixed vector of random component including sires effect and dam effect within sires; and e is the vector of residual effect.
RESULTS AND DISCUSSION
Sequence analysis of porcine FcRn gene
The porcine FcRn cDNA is 1,577 bp in length (GenBank Accession Number: AY135635) and contains a 1,077 bp open reading frame (ORF) encoding a 356-amino acid polypeptide. The 3′ end of the sequence contains a poly(A) stretch, preceded by a putative polyadenylation signal AATAAA (nucleotides 1523–1529). Blast analysis revealed that the mRNA sequence of the porcine FcRn gene has 79.4%, 66.3% and 83.9% nucleotide identity with the corresponding gene in human, mouse and cattle respectively. Comparison of predicted porcine FcRn amino acid sequence with that of human, mouse, rat and cattle indicated an identity of 77.1%, 66.3%, 64.2% and 83.9%, respectively (Figure 1). The complete porcine FcRn genomic DNA sequence spans 8,900 bp (GenBank accession number: HQ026019) and it consists of 5 introns separating 6 exons. The intron/exon organization of porcine (5 introns and 6 exons) is identical with that of the human and mouse FcRn gene. These sequence characters will provide help in further understanding of the function of porcine FcRn in antiviral defense and immune regulation.
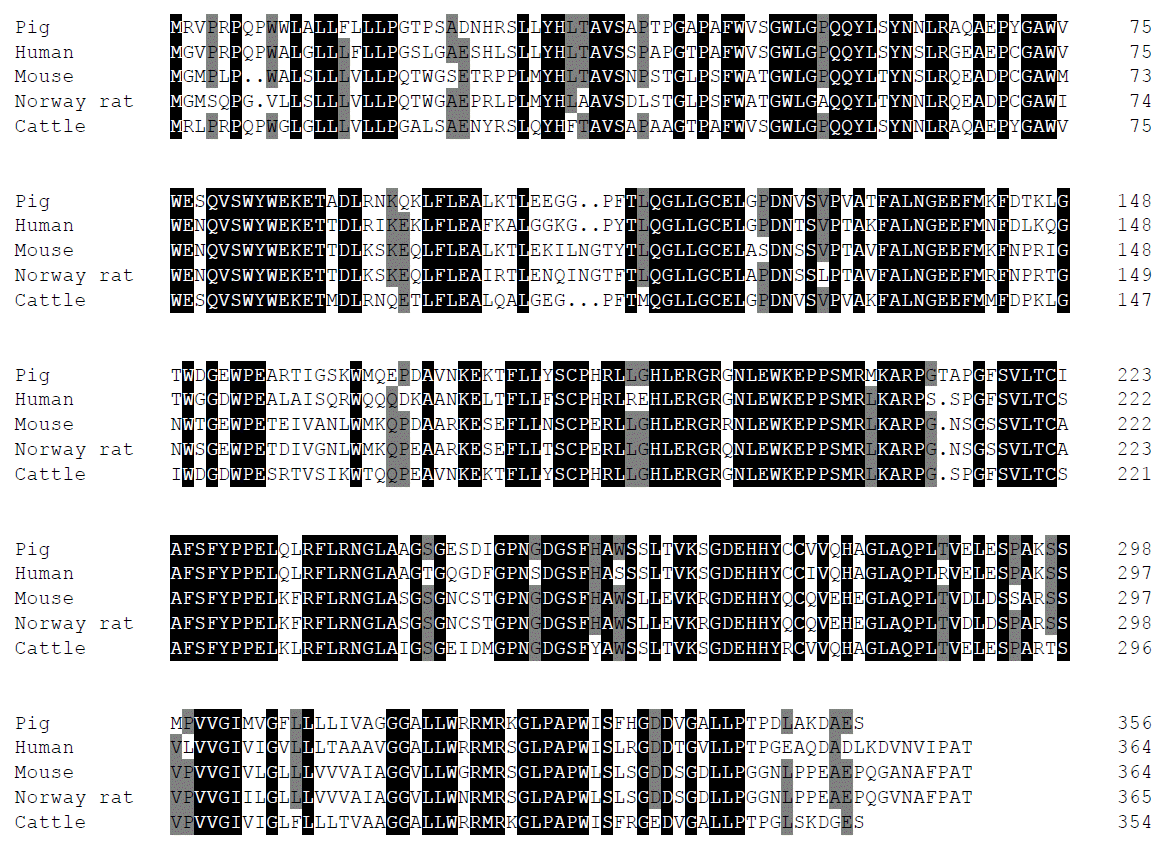
Alignment of predicted porcine FcRn amino acid sequence (Sus scrofa, NP_999362.2) with other species. The degree of homology is indicated by darker shading and justifying greater homology. Protein sequences are from GenBank (locus numbers: Human (Homo sapiens, NP_001129491.1), Mouse (Mus musculus, NP_034319.2), Norway Rat (Rattus norvegicus, NP_036986) and Cattle (Bos taurus).
Tissues expression of porcine FcRn gene
To determine tissue distribution of the porcine IRF6 gene, real time quantitative PCR (Q-PCR) was applied to ten different tissue samples. Results showed that the mRNA of FcRn was expressed in all analysis tissues (Figure 2). The higher expression level was detected in liver, spleen, thymus and lymphoid. In addition, the relatively lower expression level was detected in non-immune tissues such as muscle and brain. There may be a potential relationship between higher expression level and immune response capacity, which should be further verified in a larger sample. In some previous studies, FcRn mRNA has been detected in many tissues of adult rats, mice and humans, and FcRn is expressive in several adult tissues and in cell lines such as the vascular endothelium, professional APCs and the hepatocytes, which are consistent with our results in pigs (Israel et al., 1997; Simister et al., 1997; Brovak et al., 1998; Spiekermann et al., 2002; Shah et al., 2003; Ward et al., 2003; Mayer et al., 2004).
Polymorphism detection and association analysis of porcine FcRn gene
Comparison of sequences among three pig populations revealed one SNP (HQ026019:g.8526 C>T) in exon6 region of the porcine FcRn gene, and this SNP is a synonymous mutation (Figure 3). The genotypic and allele frequencies of this SNP in three pig populations are presented in Table 2. Results showed that genotype CC was dominant in Landrace and Large White populations, the allele C has higher frequencies than allele T. Genotype TT was only detected in Chinese indigenous breed Songliao Black population and genotype CT was a dominant genotype and allele C also had higher frequency in the Songliao Black population.

The sequencing maps of PCR fragments for exon6 region of porcine FcRn gene with the three genotypes of the SNP (HQ026019:g.8526 C>T) indicated by black arrows.
In the further association analysis, the SNP was highly significantly associated with CSFV antibody level (At d 20; At d 35) in serum (p = 0.0008 and p = 0.0001 respectively) (Table 3). The results provided a potential relationship between FcRn gene and serum CSFV antibody level. In order to further understand the effective value of different genotypes acting on serum CSFV antibody level, we implemented multiple comparison tests for genotypes of the significant SNP. The results showed that genotypes TT, CT and CC showed highly significant differences in three pig populations (p<0.01), and pigs with genotype TT have higher serum CSFV antibody levels (At d 20; At d 35) than pigs with genotypes CC and TC (Table 3).
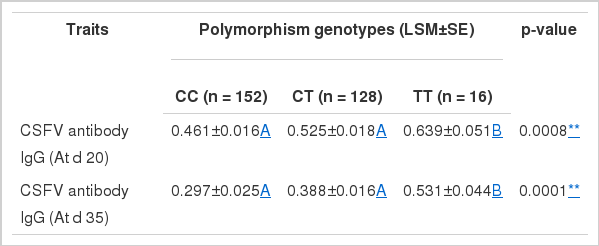
Association analysis and multiple tests of the SNP (HQ026019:g.8526 C>T) of FcRn gene with serum CSFV antibody level in three pig populations
CSFV can lead to classical swine fever (CSF), which is a highly contagious swine disease worldwide, and domestic pigs are the major natural host of this virus. CSFV could cause acute disease and mortality of fetuses, resulting in serious losses to pig industry (Dong and Chen, 2007). Thereby, improvement in the serum CSFV antibody level would help the host in defending against infection by CSF virus. Due to the important role of the FcRn gene in both perinatal IgG transport and IgG homeostatic functions, it can help in modulating the function and the serum level of IgG in the immune response._In this study, the detected SNP was not located in the promoter region and was not a functional mutation, however, synonymous SNPs have been reported that can affect protein expression by alteration or increase in the stability of the mRNA (Capon et al., 2004) and a ‘‘silent’’ polymorphism can change substrate specificity (Kimchi-Sarfaty et al., 2007). Our results showed that the SNP of FcRn gene had significant effects on serum CSFV antibody level in three pig populations and also indicated FcRn gene may be used as a genetic marker in pig disease resistance breeding.
Genetic background is one of the important factors that influence the immune capacity. In the multiple comparison tests for the SNP genotypes, genotype TT showed higher effective values than other genotypes in the serum IgG level, and it was the only detected in Chinese indigenous breed Songliao Black pig. These are interesting and can be used in the future for exploring the differences in the resistance to CSF between the Chinese indigenous pig breed and other Western commercial pig breeds. Since the number of pigs analyzed in our study was limited, therefore, further investigations are needed to confirm the relationship between the SNP with serum antibody level among other pig populations.
In summary, an 8,900 bp genomic organization with six exons and five introns of the FcRn gene was identified. The results of mRNA expression revealed that the porcine FcRn gene was widely expressed in ten analyzed tissues. One significant SNP (HQ026019:g.8526 C>T) in exon6 region of FcRn gene was also detected and association analysis results revealed that the SNP was highly significantly associated with serum anti-CSFV antibody level in three pig populations. The results indicated that FcRn gene should be regarded as a genetic marker with effects on serum anti-CSFV antibody level in pig disease resistance breeding programmes.
ACKNOWLEDGEMENTS
This work is supported by the National Major Special Project of China on New Varieties Cultivation for Transgenic Organisms (Grand No. 2009ZX08009-146B) and China Postdoctoral Science Foundation Funded Project (Grand No. 2011M500455).


