Genetic Diversity of mtDNA D-loop and Maternal Origin of Three Chinese Native Horse Breeds
Article information
Abstract
In order to protect the genetic resource of native horse breeds, the genetic diversity of mitochondrial DNA (mtDNA) D-loop of three native horse breeds in western China were investigated. Forty-three 600 bp mtDNA D-loop sequences were analyzed by PCR and sequencing techniques, 33 unique haplotypes with 70 polymorphic sites were detected in these horses, which account for 11.67% of 600 bp sequence analyzed, showing the abundant genetic diversity of the three native horse breeds in western China. The Neighbour-Joining (NJ) phylogenetic tree based on 247 bp of 43 D-loop sequences demonstrated the presence of seven major lineages (A to G), indicating that the three native horse breeds in western China originated from multiple maternal origins. Consistent with the front, the NJ phylogenetic tree based on 600 bp of mtDNA D-loop sequences of 43 Chinese western native horses and 81 sequences of six horse breeds from GenBank indicated that the three horse breeds had distributed into the seven major lineages (A to G). The structure of the phylogenic tree is often blurred because the variation in a short segment of the mitochondrial genome is often accompanied by high levels of recurrent mutations. Consequently, longer D-loop sequences are helpful in achieving a higher level of molecular resolution in horses.
INTRODUCTION
Horses have played a pivotal role in the development of human society. As an important domestic animal, the origin and history of modern domestic horses have been studied for decades in the biology and archaeology fields. At present, it is widely believed that horses were domesticated from several wild populations 4,000 to 6,000 years ago (Vila et al., 2001; Outram et al., 2009). Subsequently, they differentiated into many types after long-term domestication (Jansen et al., 2002). In history, horses differed from other domestic animals that were kept as food sources, and attained a prominent role as animals of transport and warfare, changing societies on a continent-wide scale. Literature reports the importation of horses into China from different parts of the world throughout history (Xie, 1987). As a strategic military asset, governments paid much attention to horse breeding throughout Chinese history. Some archaeological evidence and genetic data supports China as one of the earliest areas of horse domestication (Lei et al., 2009). China has rich horse genetic resources, the number of domestic horses in China was 7.639 million in 2004 (Statistic report in China, 2004). Chinese indigenous horse breeds are classified into five types based on geographical distribution and characteristics. They are Mongolian, Kazakhstan, Hequ, Southwest and Tibetan types (Chang, 2009). Four horse types are distributed in western China and still act as an important source of animal power because regions such as Guizhou, Shaanxi and Xinjiang are mountainous and pastoral. Ningqiang horse is an ancient breed. Individuals of this breed with a height at the withers less than or equal to 106 cm are called Ningqiang pony, while the other with a height greater than 106 cm are called Ningqiang horse (Hou, 1989). With the development of mechanization, the role of horse as a provider of transport and draft power is decreasing. The amount of horses declined quickly in recent years. So it is urgent to study and protect horse genetic resources.
The mtDNA is a small extra-chromosomal genome that is maternally inherited. The equine mtDNA has an approximate length of 16,660 bp and consists of a coding region which contains 37 genes and a non-coding region called the D-loop region of approximately 1,192 bp (Wolstenholme, 1992; Boore, 1999; Bowling et al., 2000). The mtDNA sequences have been widely used to study taxonomy, phylogeny, genetic structure and biological identity (Peng et al., 2007; Ji et al., 2008; Ki et al., 2010). D-loop mtDNA is an important tool to study equine diversity, phylogenetic relationships and origins, such as the Mongolian horse (Lei et al., 2009), Cheju horse (Yang et al., 2002), Debao pony (Jiang et al., 2011), Japanese horse (Kakoi et al., 2007), Tibetan horse (Xu et al., 2007), Przewalskii’s wild horse (Ishida et al., 1995) and donkey (Lei et al., 2005). The diversity and origin of the Ningqiang pony, Guizhou horse and Kazakh horse as indicated by microsatellite markers and mtDNA D-loop have been reported (Du et al., 2009; Lei et al., 2009). However, the mtDNA D-loop sequence (247 bp) studied in the literature was short and might not fully show the diversity and origin of the three horse breeds in western China. Consequently, a 600 bp mtDNA control region of 43 animals from Ningqiang pony, Guizhou and Kazakh breeds and 81 mtDNA D-loop sequences of 6 breeds from GenBank (Table 1) were used in this present study to determine the diversity and origin of the three native horse breeds in western China.
MATERIALS AND METHODS
Blood sample collection and DNA isolation
Blood samples were obtained from 43 horses of three western China native horse breeds including Ningqiang pony, Guizhou and Kazakh horse breeds and stored at −70°C. Ningqiang pony samples were collected from Ningqiang county, Hanzhong city, Shaanxi province; Guizhou horse samples came from Bijie region, Guizhou province; Kazakh horse samples were collected from Shihezi city, Xinjiang Autonomous Region. In addition, 81 mtDNA D-loop sequences from six domestic horse breeds and one donkey sequence as an outgroup were selected from GenBank (Table 1). The DNA used in PCR amplification was isolated from blood samples using the phenol-chloroform procedure.
PCR amplification and sequencing
The mtDNA D-loop sequences was amplified by the following primers: F: 5′-CGACAACAATTCACCCTCAT-3′, R: 5′-GAAGAAGGGTTGACAGATTTA-3′. Amplification was performed in a 25 μl reaction containing 50 ng of genomic DNA, 0.25 μM of each primer, 0.25 mM dNTPs, 2.5 mM MgCl2, 1.5 U Taq DNA polymerase and 10×buffer. Thermal cycling was carried out on an MJ Research 200 Thermal cycler. The procedure included predenaturation at 94°C (3 min), followed by 34 cycles of 94°C (30 s), 55°C (30 s), 72°C (90 s), and then final extension at 72°C for 10 min. The 1,100 bp PCR production was purified and two-way sequenced by ABI 3130 automated sequencer.
Statistical analysis
In this study, 43 mtDNA D-loop sequences of three horse breeds were sequenced, and aligned with the DNAMAN4.0 software (Tamura et al., 2007). Because of the number of repeats was variable within individuals (Kim et al., 1999), the repeat region was excluded and these sequences were truncated to 600 bp. Polymorphic sites, haplotype numbers, haplotype diversity (h) and nucleotide diversity (p) were calculated by DNASP 5.0 software (Rozas et al., 2003). The NJ tree of different horse breeds were constructed based on the Kimum 2-parameter model.
RESULTS
Genetic polymorphism of mtDNA D-loop region of three Chinese native horse breeds
Equine 600 bp of mtDNA D-loop sequence was examined in 43 Chinese domestic horses. The average ratio of the four nucleotides A, T, C, G was 28.3%, 27.5%, 27.0% and 17.2%, respectively.The contents of A+T exceeded G+C, showing that A+T was richer at the mtDNA D-loop region. This was in accordance with the order of nucleotide composition in the vertebrate mitochondrial genome, which is A>C>T>G with a skewed base composition and with more A+T than G+C base pairs (Asakawa et al., 1991; Ji et al., 2008). Thirty three mtDNA haplotypes were identified. 70 polymorphic sites (Figure 1) were detected, which represented 11.67% of the total mtDNA sequence analyzed (600 bp). Only one of the 70 polymorphisms represented insertion/deletion of single base pairs. The remaining were 67 transitions and two transversions, indicating a strong bias towards transitions that was common in mammalian mitochondrial evolution (Kim et al., 1999).
Thirty-three mtDNA haplotypes were detected in 43 Chinese western native horse samples (Table 2). The number of haplotypes identified in the three western China native horse breeds ranged from nine to 15. The haplotype diversity and nucleotide diversity of the mtDNA D-loop region in three Chinese horse breeds are shown in Table 2. The haplotype diversity values ranged from 0.977±0.023 in the Kazakh breed to 0.989±0.031 in the Guizhou breed. The comparing result of nucleotide diversity was consistent with that of haplotype diversity. The data indicated an abundant genetic diversity in Chinese western domestic horse breeds.
Phylogenetic analysis of different horse breeds based on mtDNA D-loop sequence
A Neighbor-Joining (NJ) phylogenetic tree was constructed based on 247 bp of mtDNA D-loop hypervariable region sequences of 43 Chinese western native horses (Figure 2). Based on the previously published seven major mtDNA lineages (A to G) in horses (Vila et al., 2001; Jansen et al., 2002; McGahern et al., 2006), the mtDNA haplotypes in Chinese three western horse breeds demonstrated the presence of these seven major lineages (A to G) (Figure 2). Ningqiang pony breed was distributed into two lineages (A, F), Guizhou breed had four lineages (A, B, D, G), and Kazakh breed had five lineages (A, C, D, E, F). Consistent with the front, when the NJ phylogenetic tree was constructed based on 600 bp of equine mtDNA D-loop sequences of 43 Chinese western native horses and 81 sequences of different horse breeds from GenBank (Table 1) it indicated that the three Chinese western horse breeds had distributed into the seven major lineages (A to G) (Figure 3). In detail, according to the NJ tree based on 600 bp of mtDNA D-loop sequences, Ningqiang pony breed was distributed into two lineages (A, F), Guizhou breed was distributed into three lineages (A, B, D, G), and Kazakh breed was distributed into four lineages (A, C, E, F). In addition, Baise breed was grouped into four lineages (A, C, D, G), Mongolian breed was grouped into four lineages (A, B, D, F), Debao pony breed was grouped into six lineages (A, B, C, D, F, G), Arabian breed was grouped into one lineage (A), Cheju breed was grouped into four lineages (A, B, C, F), Yunnan breed was grouped into five lineages (A, C, D, F, G). Among the three horse breeds and six breeds available from GenBank, lineage A was predominant. The results indicated that China western native horse breeds derived from multi-matrilineal origins and interchanged genetic material with each other widely during their evolutionary history.
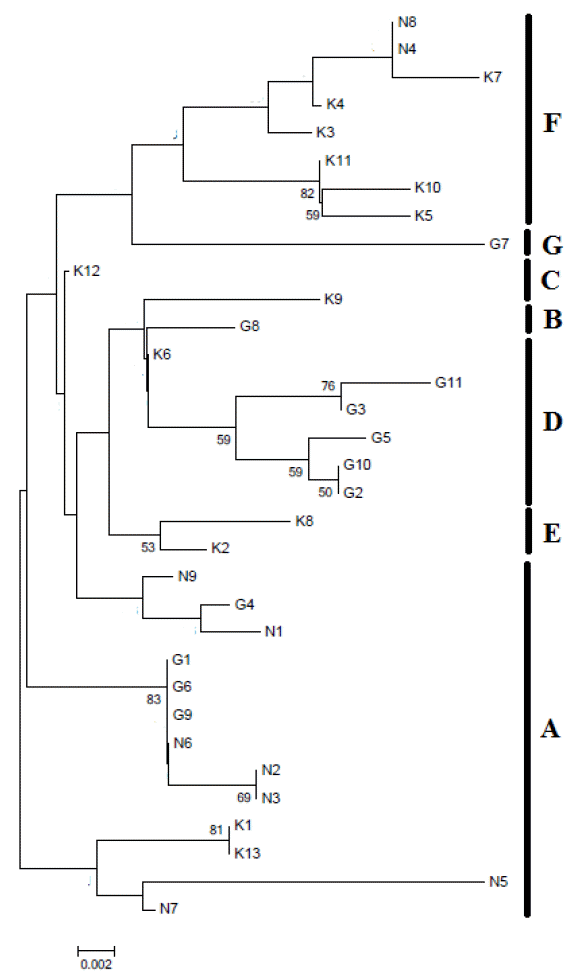
Neighbour-Joining tree based on 247 bp of mtDNA D-loop region in three Chinese western horse breeds. Bootstrap values greater than 50 in 1,000 replications are shown at the respective nodes.
DISCUSSION
In recent years, many researches have indicated the diversity and origin of different horse populations, and suggested that domestic horses were derived from several wild populations (Lister, 1998; Vila., 2001; Jansen, 2002). Wallner (2003) studied the unique nucleotide sequence of the Y chromosome, and found that Przewalskii haplotype was the ancestor. Lei et al. (2009) showed that the domestication of horses was not a single discrete event, multiple horse domestication events may have occurred later in other regions, and that China was one of these domestication regions.
We found 33 haplotypes in 43 horses from three western China native horse breeds based on 600 bp of mtDNA control region sequence, and all these haplotypes grouped into seven lineages. These three horse breeds shared A lineage, and also had their respective lineages. Nine haplotypes were identified in ten Ningqiang ponies, and were distributed into two lineages (A, F). There were 13 haplotypes in 14 Guizhou horses representing three lineages (A, B, D, G). In 19 Kazakh horses, 15 haplotypes were distributed into four lineages (A, C, E, F). These data showed that the three western China native horses possessed abundant mtDNA diversity and indicated multiple maternal origins. The results were in accordance with other results from Yunnan, Mongolian, Debao, Guanzhong and other horse breeds in China (Wang et al., 1994; McGahern et al., 2006; Li et al., 2009). Some Ningqiang ponies, Guizhou horses and Kazakh horses were clustered into A lineage, indicating lineage A is predominant. Ningqiang pony breed demonstrated only A and F lineages, indicating close genetic relationship within this breed, which was consistent with the results of Lei et al. (2009). However, in the study of Lei et al. (2009), the Ningqiang breed indicated A, B, D and F lineages, Kazakh breed indicated A, B, C, D, F and G lineages, and Guizhou breed indicated A to G lineages. The possible reason for this difference with the present study is that more samples were analyzed in the study of Lei et al. (2009). It is necessary to point out that the lineages in the study of Lei et al were based on 247 bp while in the present study 600 bp D-loop sequences were used in these three horse breeds. In many other mammalian species, the variation in this short segment of the mitochondrial genome is often accompanied by high levels of recurrent mutations, thus blurring the structure of the tree and rendering the distinction between some important ancient branches within the tree virtually impossible (Achilli et al., 2009). So, to improve mtDNA phylogeny, the longer D-loop sequences in mitochondrial genomes will be helpful to achieve the highest level of molecular resolution in horses (Achilli et al., 2012).
In this study, three typical horse breeds were selected to clarify the origin, evolutionary relationship and exchange background of western horses in China. Kazakh horse is an ancient breed, and distributed in the Chinese northwest region, while the Guizhou horse is a southwest horse breed. Ningqiang pony is a breed from the border land between northwest and southwest and possesses some characteristics of northern and southern horse populations. Many researchers claim that Ningqiang pony belongs to the Southwest type. The recent results of archaeological studies indicated that there was a chama gudao (Tea-Horse road in ancient times) in southern Shaanxi and that Ningqiang county was an important center of the Tea-Horse trade system. Combined with the geography, history and the data from genetics, we believe that Ningqiang ponies were affected by northern and southern horse populations, such as the Mongolian, Guizhou and Kazakh breeds. Our data demonstrated that Chinese native horse breeds had extensive genetic flow from breeds all over the world. The results were in accordance with the historical records that horses were continuously imported for military forces and trade during Chinese history (Xie, 1987).
ACKNOWLEDGEMENTS
The work was supported by Scientific Research Fund of Education Department of Shaanxi Province (No. 06JK328 and 07JK212).



