 |
 |
| Anim Biosci > Volume 36(9); 2023 > Article |
|
Abstract
Objective
This study was conducted to investigate polymorphisms of the melanocortin-4 receptor (MC4R) and insulin like growth factor 2 (IGF2) genes and to evaluate the growth traits affected by such polymorphisms in Thai native (Kradon) pigs.
Methods
Blood samples and productive data from 91 Kradon pigs were collected. DNA was extracted and quantified, the IGF2 and MC4R genes were amplified, and the polymerase chain reaction (PCR) produces were digested using the PCR-restriction fragment length polymorphism (PCR-RFLP) technique. Genotyping was performed, and the association between genotypes and growth traits on the birth and weaning weights were evaluated.
Results
The IGF2 intron7 g.162G>C variations in Kradon pigs were found in three genotypes: i) GG, ii) GC, and iii) CC. The GG genotype frequency was the highest followed by the GC and CC genotypes. The frequencies of the G and C alleles were 0.703 and 0.297, respectively. The MC4R genotype was found in only one genotype (GG). The IGF2 gene pattern was not associated with birth weight traits, whereas the IGF2 gene pattern was related to the weaning weight trait in Kradon pigs. Pigs with the CC and GC genotypes had higher weaning weights than ones with the GG genotype (p<0.001).
Four distinct breeds of Thai native pigs, Hainan, Rad, Puang, and Kwai, are found in Thai farms [1]. The Rad or Kradon pigs, found in the Northeast of Thailand [2], have the smallest body size among Thai native pigs and are entirely black in color, have a fine meat texture, small bones, and delicious meat [3]. Generally, they are raised in the backyards in the Northeast Thailand and are usually fed with residues of plant products found around houses and crops. Kradon pigs have long been associated with rural peoples’ culture as most of this population raises Kradon pigs for household consumption during festivals, such as New Year and Songkran. Kradon pigs are adapted to their environment and have delicious meat even after consuming low quality feeds. However, Thai native pigs are decreasing in number, due to a slow-growth rate and poor carcasses quality that has high abdominal fat. Therefore, the selection for growth and red meat production is necessary for increased production and to conserve the genetic diversity of Thai native pigs.
The insulin like growth factor 2 (IGF2) gene (or somatomedin A) is located on chromosome 2 (SSC2). This gene spans 30 kbp and consists of nine exons, eight introns, and four promoters. IGF2 exons 7, 8, and 9 encode prepro IGF2 protein [4], and exons 1–6 are non-coding [5]. The IGF2 gene is one of the potentially important genetic markers for the growth and carcass traits of pigs [6]. IGF2 concentration is associated with body weight in growing pigs [7]. The substitution at the IGF2 intron7 g.162G>C results in increased growth and body size of pigs [8,9] but a decrease in body length (BL) and back fat (BF). According our previous study [10], the association of IGF2 gene with back fat depth at 10t rib and last rib in Thai native pig was revealed. The Canadian Center for Swine Improvement (CCSI) cooperates with GENTEC, Belgium to investigate the effects of the IGF2 gene in a Canadian wild pig population. It was found that the IGF2 gene controls red meat production in wild pigs. Thus, the IGF2 gene can be used to both increase and decrease the amount of red meat in wild pigs. This report will be useful in creating commercial wild pig breeds, which will be capable of producing wild pigs that are uniform in terms of red meat production. The IGF2 gene might be used to select Thai native pigs to improve growth performance.
The melanocortin-4 receptor (MC4R) gene in pigs is primarily responsible for energy homeostasis. It is located on chromosome 1 and has a total sequence length of 18 kp with 1 exon. At position c.746G>A (Asp298Asn), the transition from aspartic (Asp) to asparagine (Asn) was found to be associated with an increase in growth rate (average daily gain [ADG]) and feed intake (FI) in pigs [11–13]. The pigs without the MC4R mutation showed thinner BF traits, lower ADG, and less FI. The mutation of the MC4R gene was used as a DNA marker to improve the growth traits of pigs.
Hence, the IGF2 and MC4R genes were suitable candidate genes for growth trait studies. The objectives of this study were to investigate the polymorphisms of the IGF2 and MC4R genes and evaluate whether these polymorphisms affect the growth traits in Kradon pigs.
This experiment was reviewed and approved by the Institutional Animal Care and Use Committee of Rajamangala University of Technology Isan (No.48/2564).
A random population from a total of 91 blood samples of Kradon pigs (39 castrated males and 52 females) were collected from the swine farm unit of the Department of Animal Science, Faculty of Natural Resources, Rajamangala University of Technology Isan, Sakon Nakhon Campus. All pigs were raised under an open-house system in cement pens of 2×4 m and were handled with identical procedures. The pigs received clean water from a nipple drinker and were fed with a diet of not less than 20% crude protein (CP). Their birth and weaning weights were recorded. Blood samples were collected from the jugular vein by needles No.18G×1 in for a volume of 5 to 10 mL of collected blood and added to a 15 mL tube containing 0.5 M ethylenediaminetetraacetic acid. These samples were centrifuged at 4,200 rpm for 5 min to precipitate the white blood cells and then stored at −4°C until they were used for DNA extraction.
The buffy coat or white blood cells was prepared from 50 μL of whole blood added to 1.5 mL microtubes after which 1,000 μL 0.9% NaCl was added to the tubes, and then mixed and vortexed for 5 to 10 s. After centrifugation at 10,000 rpm for 5 min, the supernatant was discarded. Genomic DNA was extracted by a guanidine hydrochloride protocol modified procedure developed by Goodwin et al [14]. Fifty microliters of white blood cells were divided into 1.5 mL microtubes, and then 70 μL 20% SDS, 50 μL 7.5 M Na-acetate, 25 μL 1% Proteinase K, and 625 μL 5 M Guanidine HCl were added to the tubes and mixed by flipping the microtubes up and down 2 to 3 times and vortexed for 5 to 10 s. They were then incubated overnight at 65°C in a waterbath. These microtubes were centrifuged at 10,000 rpm for 5 min after which 500 μL of supernatant was aspirated into the new tube. Five-hundred microliters of cold absolute isopropanol was then added to this mixture, The microtubes were flipped up and down, and DNA precipitation was noted. The microtubes were then centrifugated at 10,000 rpm for 5 min after which the supernatant was discarded while the DNA precipitate was isolated. The DNA was washed by adding 500 μL of 75% ethanol and then centrifuged at 10,000 rpm for 1 min after which the supernatant was discarded twice, while the DNA was isolated. The DNA was dried at room temperature for 30 to 60 min after which 25 μL of TE buffer was added, and the DNA was incubated in a waterbath at 37°C overnight to dissolve the DNA. One microliter of the DNA sample was used to determine the DNA quality using a NanoDrop2000 spectrophotometer (Nano Drop Thermo Scientific, Wilmington, DE, USA) at 260 and 280 nm wavelengths. The DNA concentration was adjusted to 50 ng/μL and stored at −20°C for use in subsequent amplification.
The primers for the IGF2 and MC4R genes were designed from published sequences. The IGF2 primer sequence has been reported by Vykoukalová et al [15] and the MC4R primer sequence was reported by Dvorakova et al [16]. The IGF2 and MC4R fragments were amplified using polymerase chain reaction (PCR), 10 μL contained 4.1 μL distill water, 1 μL dNTP (1 mM; Fermentas, Hanover, MD, USA), 0.8 μL MgCl2 (25 mM), 1 μL 10× PCR buffer, 1 μL of 3 μM each primer (forward and reverse primer), 0.1 μL 5 U Taq DNA polymerase (MBI Fermentas, USA), and 1 μL 50 ng genomics DNA. Primers and are listed in Table 1. A T-Professional Standard Thermo cycler (Biometra GmbH, Gottingen, Germany) was used for amplification under specific conditions: i) 95°C for 3 min, ii) 30 cycles at 95°C for 30 s, iii) annealing for 30 s and 72°C for 45 s, and iv) a final extension at 72°C for 5 min. The products were evaluated using a 2% agarose gel electrophoresis.
The PCR products of IGF2 and MC4R genes were digested in a total volume of 10 μL, containing 6.7 μL of distilled water, 2 μL PCR products, 1 μL cut smart buffer, and 0.3 μL each of restriction enzymes, and the reactions were digested overnight at 37°C for BcnI, and 65°C for TaqI (New England Biolabs, Ipswich, MA, USA). The digested PCR products of IGF2 and MC4R were separated on 2% agarose gel electrophoresis at 100 V for 30 min, and RFLP fragments were visualized using Gel Star (Lonza, Rockland, ME, USA). The IGF2 gene pattern were three genotypes pattern with GG, GC, and CC. GG genotype DNA banding sizes were 308 and 28 bp, GC genotypes were 308, 208, 100, and 28 bp, and CC genotype pattern were 208, 100, and 28 bp. Additionally, MC4R genotyping with three patterns demonstrated the presence of GG, AG, and AA. The GG pattern was represented with 156 and 70 bp, GA genotype was 226, 156 and 70 bp, and AA genotype was 226 bp.
The genotype patterns of IGF2 and MC4R genes were used for calculating the gene and genotype frequencies, observed heterozygosity (HO), expected heterozygosity (HE), and unbiased expected heterozygosity (HU) following Nei’s method [17]. The polymorphic information content (PIC) was assessed according to the method by Botstein et al [18]. The Hardy–Weinberg Equilibrium of the IGF2 and M4CR genes was tested using the GENEPOP software (version 4.2) available online: http://genepop.curtin.edu.au/. The investigation of the association between the polymorphism of genotype pattern and both birth and weaning weight traits was carried out with the general linear model procedure using the SAS procedure (Statistical Analysis System 9.0; SAS Institute, Cary, NC, USA) by the following statistical linear model for birth weight trait according to Yijk = μ+Sexi+Dj+Gk+ɛijk, and weaning weight trait according to Yijk = μ+Sexi+Dj+Wk+Gl+ɛijk in which Yijk represents the phenotypic values; μ is the overall mean; Sexi represents the sex effect; Dj is the maternal effect; Wk is the covariate of birth weight effect; Gk and Gl are genotype effects; and ɛijk represents the random error.
Genotype patterns of the genes were studied from the amplified gene fragments that were digested with the restriction enzymes, and it was found that the size of the IGF2 and MC4R gene fragments matched the size of the reference reports. IGF2 gene variations in Kradon pigs found with the three genotypes were GG, GC, and CC (Figure 1A). The polymorphism of MC4R was found in only the GG genotype with fragment sizes of 156 and 70 bp (Figure 1B).
Our study of polymorphism of the IGF2 gene analyzed the allele and genotype frequencies, and it was found that the GG genotype frequency was the highest (0.516) followed by the GC genotype frequency (0.374), and that the CC genotype frequency was the lowest (0.110) as shown in Table 2 with an allele frequency G and C of 0.703 and 0.297, respectively.
The HWE analysis is reported in Table 2. The results showed the IGF2 genotype followed the null hypothesis that stated that Kradon pigs might not be affected by gene forces because this initial population was maintained in a relatively closed area without a systematic breeding program. Thus, migration and selection factors may not significantly affect the HWE of the IGF2 gene. In contrast, MC4R gene could not undergo HWE analysis because it is a non-diverse allele with only GG genotype.
An evaluation of the MC4R gene was not feasible because only one genotype pattern was found. Therefore, the association between the genotype pattern and both their birth and weaning weight traits could not be analyzed. However, the association of IGF2 with birth and weaning weight traits was analyzed. The results of the study found that the genotype pattern was not associated with birth weight traits (p>0.05). Regardless of the genotype of the Thai native pigs, no influence on the birth weight of piglets was found. However, a correlation between the IGF2 gene pattern and weaning weight traits was found (Table 3). Pigs with the CC genotype had the highest weaning weight, and pigs with the GG genotype had the lowest (p<0.001).
The genotypic pattern of the IGF2 gene, that is the CC, GC, and GG genotypes, found in the Thai native pig is the same as in commercial pig breeds such as Large White, Landrace, Duroc, Pietrain, and Yorkshire [19,20]. The Thai native pigs had GG genotype frequencies (0.516) higher than the GC and CC genotypes (0.374 and 0.110, respectively), and G allele frequencies (0.703) higher than C allele frequencies (0.297). This report was consistent with the report of Klomtong et al [19] who mentioned that the genotypic frequencies of the IGF2 genes with GG, GC, and CC genotypes were 0.570, 0.270, and 0.160, respectively. In contrast, with regard to the genotype frequency of the IGF2 gene in commercial pigs, the CC genotype has a higher genotype frequency than the GG genotype [15,19]. It can be concluded that the genetic structure of the IGF2 gene is different in Thai native pigs versus commercial pigs. This difference may be an important factor for Thai native pigs to have lower growth rates than commercial pigs. Therefore, if the genetic structure of the IGF2 gene were manipulated in the Kradon pigs so that it has a higher CC genotype, it may help the Thai native pigs have a better growth rate and an improved carcass quality than the present ones.
In this study, IGF2 gene variations were found to be related to the weaning weight characteristics of the Kradon pigs, which may be caused by hepatic IGF2 expression in the pigs. Clark et al [21] delineated the effects of the IGF2 mutation on the expression of myogenic gene during prenatal and postnatal growth. Male and female offspring were evaluated at birth (0 d) and weaning (21 d), and it was found that at 21 d hepatic, the IGF2 expression was greater (p = 0.01). The increased IGF2 expression may contribute to an increase in the muscle mass of pigs [22]. The partial knockout of zinc finger BED-type containing 6 (ZBED6) could affect the secretion of IGF2 in pig liver [23]. The interaction between the ZBED6 and the IGF2 locus plays a prominent role in up-regulating postnatal growth of skeletal muscle and internal organs; kidney, liver, and heart [24]. Reports of the IGF2 gene expression and its effect on fatty acid composition in different pig genetic backgrounds are available. Twenty-five percent of the gene expression indicated significant associations between IGF2 polymorphism and arachidonic, hexadecenoic, oleic, linoleic, and α-linoleic fatty acids and the monounsaturated fatty acid/polyunsaturated fatty acid (MUFA/PUFA) ratio measured in BF [25].
The IGF2 gene has also been reported to be associated with economically important traits of various pig breeds, such as the genotypic pattern of the IGF2 gene in increased red meat and fat thickness in pigs [8,20]. Braunschweig [26] reported that the IGF2 gene is expressed in organs, such as the brain, liver, muscles, and kidneys. Therefore, it is possible that the IGF2 gene associations may be found at all stages of the maturation of pigs during the fetal and postnatal stages [27]. This association may depend on the breed of the pigs or the size of the population used in the study. Duroc pigs, especially, have high frequencies of alleles at the IGF2 intron 3 g.3072G>A loci that favor lean production and stress resistance [28].
The polymorphisms of the IGF2 gene have the potential to be used for marker-assisted selection of growth traits in Kradon pigs. The selection and mating of animals with CC and GC genotypes can help improve the growth performance of Kradon pigs. However, to more precisely assess the effects of IGF2 polymorphism on growth and meat production, it would be necessary to increase the population size.
This work differs from that reported by Chaweewan et al [29] who reported that the MC4R genotypes, AA, GA and GG genotypes were found in the hybrid pig breeds, Pak Chong 1–5. This difference may be due to a decrease in the population size of Kradon pigs in Thailand, which may be a factor affecting the gene diversity of the MC4R gene in Kradon pigs when compared with Large White, Landrace, Duroc, Pietrain, and other pig breeds, developed by the Department of Livestock Development of Thailand such as Pak Chong 1–5. In this study, the number of MC4R gene variants differed from those reported by Fan et al [30] and Szyndler-Nędza et al [31] who found three MC4R genotypes in pigs. Piórkowska et al [32] suggested that the A allele should be selected to produce improvements in litter size and fertility in the dam line and to increase the frequency of the G allele in the sire line to obtain pigs with high red meat content.
The MC4R gene is not only related to the growth characteristics of pigs but is also associated with an increase in fat thickness [33] and feed intake in pigs [12]. The missense mutation of the MC4R gene with alternating G and A bases resulted in the amino acid at position 298 shifting from the aspartic (Asp) codon, GAU, and GAC to an asparagine (Asn) coded AAU and AAC codon whose A allele was found to correlate with feed intake, whereas the G allele was associated with high red meat content in commercial and hybrid pigs [29]. However, in this study, only a single genotypic pattern of the MC4R gene was found in the Kradon pig. Therefore, to study the association of MC4R genes with economically important traits of Kradon pigs, the number of samples should be increased to have a variety of genes.
Notes
ACKNOWLEDGMENTS
The research was supported by animal blood samples from the Rajamangala University of Technology Isan, Sakon Nakon Campus. The authors are grateful to the Breeding and Omics Laboratory, Northeastern Science Park (Khon Kaen), Thailand, and especially thank the Animal Biotechnology Laboratory, Animal Production and Management Division, Faculty of Natural Resources, Prince of Songkla University, Songkla, Thailand.
Figure 1
PCR-RFLP patterns: (A) IGF2/BcnI; (B) MC4R/TaqI genes in Thai Kradon pigs. PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism; IGF2, insulin like growth factor 2; MC4R, melanocortin-4 receptor. M = 100 bp DNA ladder; PCR = PCR product; GG, GC, and CC = genotypes.
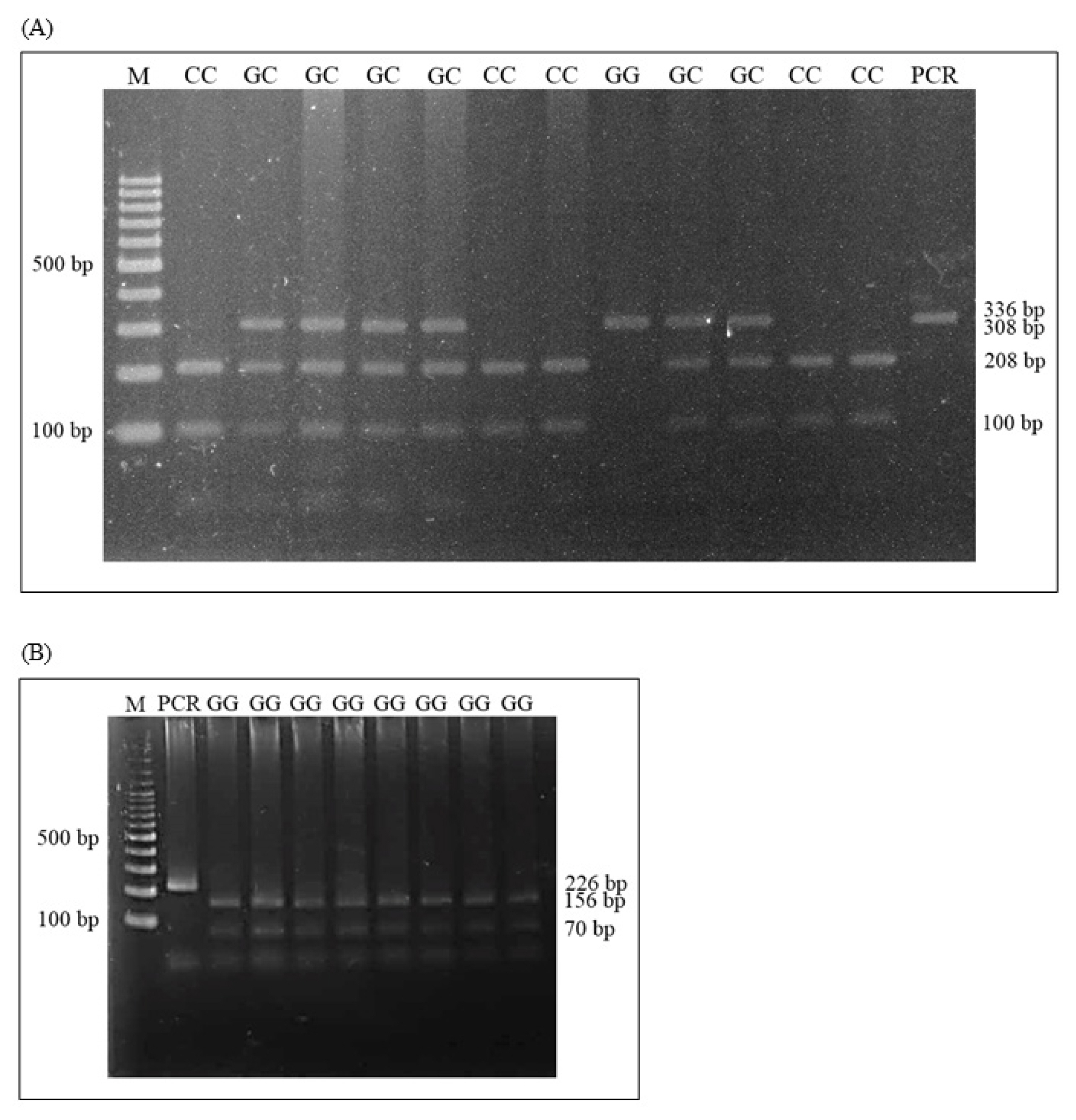
Table 1
Accession number, primer sequences, Tm and PCR product sizes of IGF2 and MC4R markers
Table 2
Genotype and allele frequencies, HO, HE, HU, PIC, and chi-square test (x2) of IGF2 and MC4R genes in Thai Kradon pigs
REFERENCES
1. Falvey L. Research on native pigs in Thailand. Rome, Italy: World Animal Review; 1982.
2. Vasupen K. Nutrition studies in native, Thai Kadon pig [Ph.D. Thesis]. Utrecht, The Netherlands: Department of Nutrition, Faculty of Veterinary Medicine, Utrecht University; 2007.
3. Rattanronchart S. Thai native pigs [internet]. c2021. [cited 2021 Dec 9]. Available from: https://www.angelfire.com
4. Amarger V, Nguyen M, Van Laere AS, et al. Comparative sequence analysis of the INS-IGF2-H19 gene cluster in pigs. Mamm Genome 2002; 13:388–98.
https://doi.org/10.1007/s00335-001-3059-x


5. O’ Dell SD, Day INM. Molecules in focus Insulin-like growth factor II (IGF-II). Int J Biochem Cell Biol 1998; 30:767–71.
https://doi.org/10.1016/S1357-2725(98)00048-X


6. Kostyunina OV, Kramarenko SS, Svezhentseva NA, Sizareva EI, Zinovieva NA. The association of IGF2 with productive traits of pigs of large white breed in the aspect of sexual differentiation. Agric Biol 2015; 50:736–45.
https://doi.org/10.15389/agrobiology.2015.6.736eng

7. Lamberson WR, Sterle JA, Matteri RL. Relationships of serum insulin-like growth factor II concentrations to growth, compositional, and reproductive traits of swine. J Anim Sci 1996; 74:17536
https://doi.org/10.2527/1996.7481753x

8. Burgos C, Galve A, Moreno C, et al. The effects of two alleles of IGF2 on fat content in pig carcasses and pork. Meat Sci 2012; 90:309–13.
https://doi.org/10.1016/j.meatsci.2011.07.016


9. Thengpimol P, Suwanasopee T, Mekchay S, Koonawootrittriron S. Association of the Insulin-like Growth Factor II Gene (IGF-II) with growth and body conformation traits in a commercial swine population. Khon Kaen AGR J 2009; 37:319–30.
10. Ampaporn K, Parnprasert P, Sanklong Ch, Duangjinda M. The effects of IGF-II gene on growth trait and carcass quality in Kradon pigs. Khon Kaen AGR J 2018; 46:533–42.
11. Kim KS, Larsen N, Short T, Plastow G, Rothschild MF. A missense variant of the porcine melanocortin-4 receptor (MC4R) gene is associated with fatness, growth, and feed intake traits. Mamm Genome 2000; 11:131–5.
https://doi.org/10.1007/s003350010025


12. Meidtner K, Wermter AK, Hinney A, Remschmidt H, Hebebrand J, Fries R. Association of the melanocortin 4 receptor with feed intake and daily gain in F2 Mangalitsa x Piétrain pigs. Anim Genet 2006; 37:245–7.
https://doi.org/10.1111/j.1365-2052.2006.01414.x


13. Bruun CS, Jørgensen CB, Nielsen VH, Andersson L, Fredholm M. Evaluation of the porcine melanocortin 4 receptor (MC4R) gene as a positional candidate for a fatness QTL in a cross between Landrace and Hampshire. Anim Genet 2006; 37:359–62.
https://doi.org/10.1111/j.13652052.2006.01488.x


14. Goodwin W, Linacre A, Hadi S. An introduction to forensic genetics. Chicheste, UK: John Wiley & Sons Ltd; 2007.
15. Vykoukalová Z, Knoll A, Dvorák J, Cepica S. New SNPs in the IGF2 gene and association between this gene and backfat thickness and lean meat content in Large White pigs. J Anim Breed Genet 2006; 123:204–7.
https://doi.org/10.1111/j.1439-0388.2006.00580.x


16. Dvorakova V, Stupka R, Sprysl M, et al. Effect of missense mutation Asp298Asn in MC4R on growth and fatness traits in commercial pig crosses in the Czech Republic. Czech J Anim Sci 2011; 56:176–80.
https://doi.org/10.17221/1305-CJAS

17. Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978; 89:583–90.
https://doi.org/10.1093/genetics/89.3.583



18. Botstein D, White RL, Skolnick M, Davis RW. Construction of genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 1980; 32:314–31.


19. Klomtong P, Chaweewan K, Phasuk Y, Duangjinda M. MC1R, KIT, IGF2, and NR6A1 as markers for genetic differentiation in Thai native, wild boars, and Duroc and Chinese Meishan pigs. Genet Mol Res 2015; 14:12723–32.
https://doi.org/10.4238/2015.October.19.16


20. Oczkowicz M, Tyra M, Ropka-Molik K, Mucha A, Žukowski K. Effect of IGF2 of intron3-g.3072G>A on intramuscular fat (IMF) content in pigs raise in Poland. Livest Sci 2012; 149:301–4.
https://doi.org/10.1016/j.livsci.2012.06.021

21. Clark DL, Clark DI, Beever JE, Dilger AC. Increased prenatal IGF2 expression due to the porcine IGF2 intron3-G3072A mutation may be responsible for increased muscle mass. J Anim Sci 2015; 93:2546–58.
https://doi.org/10.2527/jas.2014-8389


22. Aslan O, Hamill RM, Davey G, et al. Variation in the IGF2 gene promoter region is associated with intramuscular fat content in porcine skeletal muscle. Mol Biol Rep 2012; 39:4101–10.
https://doi.org/10.1007/s11033-011-1192-5


23. Zhao H, Wu M, Liu S, et al. Liver expression of IGF2 and related proteins in ZBED6 gene-edited pig by RNA-seq. Animals 2020; 10:2184
https://doi.org/10.3390/ani10112184



24. Younis S, Schönke M, Massart J, et al. The ZBED6–IGF2 axis has a major effect on growth of skeletal muscle and internal organs in placental mammals. Proc Natl Acad Sci 2018; 115:e2048–57.
https://doi.org/10.1073/pnas.1719278115



25. Criado-Mesas L, Ballester M, Crespo-Piazuelo D, et al. Analysis of porcine IGF2 gene expression in adipose tissue and its effect on fatty acid composition. PLoS ONE 2019; 14:e0220708
https://doi.org/10.1371/journal.pone.0220708



26. Braunschweig MH. Biallelic transcription of the porcine IGF2R gene. Gene 2012; 500:181–5.
https://doi.org/10.1016/j.gene.2012.03.059


27. Xiang G, Ren J, Hai T, et al. Editing porcine IGF2 regulatory element improved meat production in Chinese Bama pigs. Cell Mol Life Sci 2018; 75:4619–28.
https://doi.org/10.1007/s00018-018-2917-6


28. Ruan GR, Xiang YY, Fan Y, et al. Genetic variation at RYR1, IGF2, FUT1, MUC13, and KPL2 mutations affecting production traits in Chinese commercial pig breeds. Czech J Anim Sci 2013; 58:65–70.

29. Chaweewan K, Srisuriya V, Jumparat V, Nakavisut S. Association of the melanocortin-4 receptor (MC4R) with economic traits in pigs. Khon Kaen AGR J 2012; 40:Suppl 2343–50.
30. Fan B, Onteru SK, Plastow GS, Rothschild MF. Detailed characterization of the porcine MC4R gene in relation to fatness and growth. Anim Genet 2009; 40:401–9.
https://doi.org/10.1111/j.1365-2052.2009.01853.x


31. Szyndler-Nędza M, Tyra M, Blicharski T, Piórkowska K. Effect of mutation in MC4R gene on carcass quality in Pulawska pig included in conservation breeding programme. Anim Sci Pap Rep 2010; 28:37–45.
32. Piórkowska K, Tyra M, Rogoz M, Ropka-Molik K, Oczkowicz M, Rózyck M. Association of the melanocortin-4 receptor (MC4R) with feed intake, growth, fatness and carcass composition in pigs raised in Poland. Meat Sci 2010; 85:297–301.
https://doi.org/10.1016/j.meatsci.2010.01.017


33. Van den MK, Stinckens A, Claeys E, et al. The Asp298Asn missense mutation in the porcine melanocortin-4 receptor (MC4R) gene can be used to affect growth and carcass traits without an effect on meat quality. Animal 2007; 1:1089–98.
https://doi.org/10.1017/S1751731107000456








 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print





