Characterization of porcine cytokine inducible SH2-containing protein gene and its association with piglet diarrhea traits
Article information
Abstract
Objective
The cytokine inducible SH2-containing protein (CISH), which might play a role in porcine intestine immune responses, was one of the promising candidate genes for piglet anti-disease traits. An experiment was conducted to characterize the porcine CISH (pCISH) gene and to evaluate its genetic effects on pig anti-disease breeding.
Methods
Both reverse transcription polymerase chain reaction (RT-PCR) and PCR were performed to obtain the sequence of pCISH gene. A pEGFP-C1-CISH vector was constructed and transfected into PK-15 cells to analysis the distribution of pCISH. The sequences of individuals were compared with each other to find the polymorphisms in pCISH gene. The association analysis was performed in Min pigs and Landrace pigs to evaluate the genetic effects on piglet diarrhea traits.
Results
In the present research, the coding sequence and genomic sequence of pCISH gene was obtained. Porcine CISH was mainly localized in cytoplasm. TaqI and HaeIII PCR restriction fragment length polymorphism (RFLP) assays were established to detect single nucleotide polymorphisms (SNPs); A-1575G in promoter region and A2497C in Intron1, respectively. Association studies indicated that SNP A-1575G was significantly associated with diarrhea index of Min piglets (p<0.05) and SNP A2497C was significantly associated with the diarrhea trait of both Min pig and Landrace piglets (p<0.05).
Conclusion
This study suggested that the pCISH gene might be a novel candidate gene for pig anti-disease traits, and further studies are needed to confirm the results of this preliminary research.
INTRODUCTION
The inflammation cytokine response is an important feature of the host’s immune response to infection caused by diverse pathogens, but excessive inflammatory response will be harmful to host. The control of inflammatory response is negatively mediated in part from the suppressor effect of cytokine signaling (SOCS) proteins through their actions on specific signaling pathways such as Janus kinase 2 (JAK2) signal transducer and activator of transcription 5 (STAT5) and nuclear factor-k-gene binding pathways [1–2]. Cytokine inducible SH2-containing protein (CISH) was the first member of the SOCS family to be described. It was initially isolated as an immediate early gene induced by granulocyte-macrophage colony stimulating factor, interleukin (IL)-2, IL-3 and erythropoietin in hematopoietic cells [3]. Then, CISH was identified as a target gene of the JAK2-STAT5 pathway [4]. CISH negatively modulates STAT5 activation by binding to the phosphorylated tyrosine residues of cytokine receptors and thereby inhibits the downstream cytokine signaling [4–6]. In addition, accumulating evidence supports the important regulatory role of CISH in the T-cell proliferation and survival in response to infection [7–9], in the type 1 dendritic cell (DC) development and DC-mediated cytotoxic T lymphocyte activation [10].
Given the central role in controlling signaling of cytokines, in particular IL-2 [11], CISH was selected as a candidate gene of human disease. Recent reports showed that the variants of human CISH were associated with susceptibility to diseases caused by diverse infectious pathogens, such as bacteremia [12], malaria [12], tuberculosis [12–13], and persistent hepatitis B [14].
In modern pig production, the large-scale intensive systems make piglets susceptible to infection by various pathogens [15]. Given the significant effects of CISH gene on human disease, in this research, we characterized the CISH gene in pigs, determined its subcellular localization, investigated genetic variation and performed association analysis with piglet diarrhea traits to evaluate its effects on pig breeding.
MATERIALS AND METHODS
Animals and data collection
In the autumn of 2013, 327 Min piglets and 203 Landrance piglets born in the same period were selected as experiment animals on the Lanxi conservation farm (Lanxi, Heilongjiang, China). All these piglets were raised with the same fodder, feeding conditions, and disease control during the experiment period. From birth to 35 days, piglet diarrhea was recorded twice per day (6:00 am and 2:00 pm) and was assigned a daily score based on a standard evaluation of symptom traits as follows: 0, normal, manure solid feces; 1, slight diarrhea, manure soft and loose feces; 2, moderate diarrhea, manure semi-liquid feces; and 3, severe diarrhea manure liquid and unformed feces [16]. Diarrhea index was the summation of diarrhea score during experiment period.
Genomic DNA was extracted from the ear tissues samples using DNA Extraction Kit (Sangon, Shanghai, China) and stored at −20°C.
Standard polymerase chain reaction reactions and sequencing
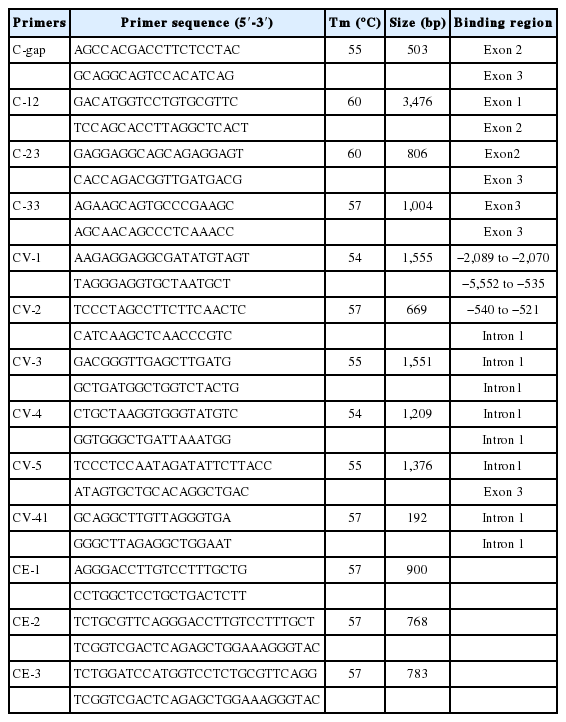
Primer pairs were designed using Primer 5.0 and then synthesized commercially (Sangon, China). The polymerase chain reaction (PCR) reaction was 25 μL containing 100 ng templates, 0.5 μM of each primer, 0.5 mM dNTPs, 2.0 U Taq DNA polymerase and 2.5 μL buffer (TaKaRa, Tokyo, Japan). PCR conditions as follows: 5 min at 94°C, 35 cycles of 45 s at 94°C, 45 s at the annealing temperature (Table 1), then 90 s at 72°C, and finally 72°C for 10 min. The PCR products were analyzed by 1.5% agarose gel electrophoresis, purified using a Gel Extraction Kit (Tiangen Biotech, Beijing, China), cloned into the pMD18-T vector (TaKaRa, Japan) and sequenced commercially (Sangon, China).
Isolation and characterization of the cytokine inducible SH2-containing protein
Total RNA of spleen tissues was isolated using Trizol Reagent (Invitrogen, Carlsbad, CA, USA). Reverse transcription (RT) was performed using the TransSript First-Strand cDNA Synthesis SuperMix (TaKaRa, Japan). Porcine expressed sequence tags (ESTs) sequences (GenBank: BE014034, DY425421, DY421524) homologous to the human orthologous genes (GenBank: NM_145071) were selected and two ESTs (DY425421 and DY421524) were assembled into a contig sequence. Primer pairs C-gap (Table 1) was used to amplify the gap sequence between EST (BE014034) and the contig. Three pairs of primers (Table 1: C-12, C-23, and C-33) were used to amplify the introns and exons of cytokine inducible SH2-containing protein (pCISH). Program Seqman (DNAstar, Madison, WI, USA) was used to identify the open reading frame (ORF) and deduce the amino acid sequences. The Sequences of CISH from different species were aligned using the Clustalw 2.0 (www.ebi.ac.uk/Tools/clustalw2/). Then the phylogenetic trees of this gene were reconstructed by the neighbor-joining (NJ) method of MEGA v4.1 program and the bootstrap tests were performed with 1,000 bootstrap replications.
Intracellular distribution of the pCISH
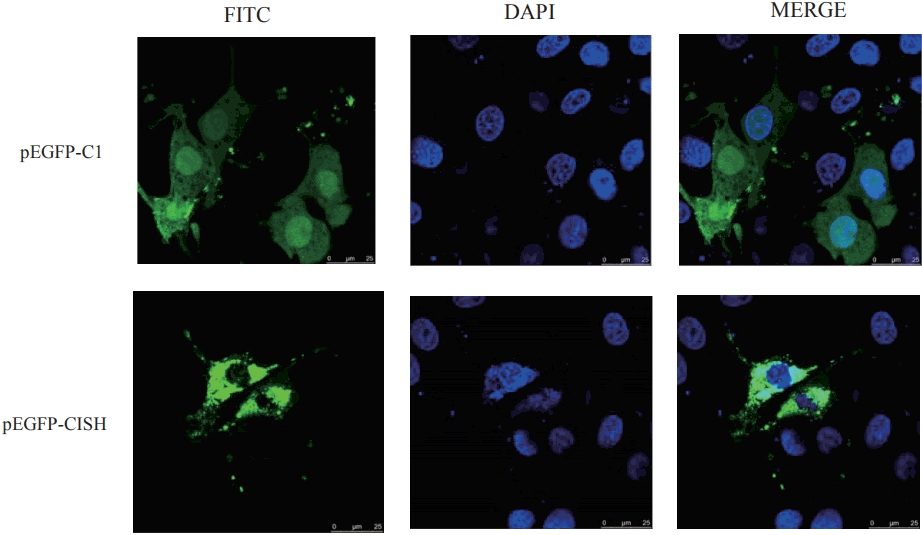
To obtain the coding sequence of pCISH gene, three pairs of primers (CE-1, CE-2, and CE-3) (Table 1) were used in the amplification and the final products were cloned into pMD18-T vectors (TaKaRa, Japan). Then, the correct insert was subcloned into the BglII-SalI site of the pEGFP-C1 vector (BD Biosciences Clontech, San Francisco, CA, USA) to construct a pEGFP-C1-CISH vector which was verified by sequencing and restriction endonuclease digestions.
PK15 cells were cultured using DMEM supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY, USA), 4 mM glutamine, 100 U/mL penicillin, and 0.1 mg/mL streptomycin under air containing 5% CO2 at 37°C. Then these cells were seeded at a density of 5×105/mL and transfected with pEGFP-C1-CISH constructs (2 μg of plasmid DNA) using Fugene HD transfection reagent (Roche, Mannheim, Germany). After 20 to 24 hours, the cells were fixed with growth medium containing 3.7% formaldehyde, incubated with 10 μM Hoechst33258. At last, the slides were mounted, sealed and analyzed by LeicaDMI 6000B Confocal Microscope (Leica, Solms, Germany).
Polymorphism identification and genotyping with polymerase chain reaction-restriction fragment length polymorphism
Genetic variation of the pCISH were analyzed by sequencing PCR products amplified by primer pairs CV-1, CV-2, CV-3, CV-4, and CV-5 (Table 1), and all these primers were partially overlapped. The PCR products from Minzhu, Landrace, and Largewhite pig breeds were purified, sequenced and then compared with each other using Clustalw 2.0. Two PCR-restriction fragment length polymorphism (RFLP) assays were designed to genotype the selected single nucleotide polymorphisms (SNPs) using primer pairs CV-1, CV-41 (Table 1) and restriction enzymes TaqI (TaKaRa, Japan), HaeIII (TaKaRa, Japan), respectively. For the PCR-RFLP profile, 8.5 μL of PCR products amplified by primer pairs CV-1 or CV-41 were digested with 5 U of TaqI or HaeIII (TaKaRa, Japan) for 5 h at 65°C or 37°C, and then separated on a 1.5% agarose gel. Genotyping work was performed in both Min pigs and Landrace population for further association analysis between pCISH and diarrhea index trait.
Statistical analysis
In this study, we used diarrhea index to evaluate the piglet diarrhea. It was analyzed as discrete data using the SAS GENMOD procedure. In this model, the dependent variable is diarrhea index, divided into 23 classes according to the summation of diarrhea score during experiment period. The independent variables are breed and genotype effect.
RESULTS
Isolation and characterization of pCISH
A 503 bp sequence from porcine spleen cDNA was obtained using primer C-gap. Together with porcine ESTs (BE014034, DY425421, DY421524), all these fragments were assembled into a 1,913 bp contig cDNA sequence of pCISH. The ORF of pCISH is 765 bp (GenBank: KM507873), which encodes a protein of 254 AA. Homology comparison showed that pCISH is similar to human, mouse, rat and cattle CISH (90% to 93%). Phylogenetic tree analysis indicated that pig and cattle homologs fell into the same cluster (Figure 1). Simple modular architecture research tool (SMART, http://smart.embl-heidelberg.de/) analysis showed that pCISH contained a SH2 domain (80–169AA), a SOCS domain (210–251AA) and a SOCS_box domain (216–250AA).
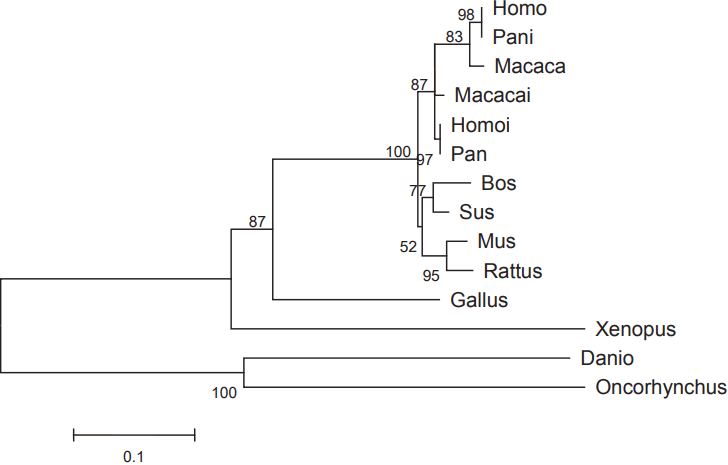
Phylogenetic tree of pCISH based on the amino acid sequence of CISH from different species. The scale bar is 0.1. The accession number of the CISH gene are as follow: Human, NM_013324, NM_145071; Rhesus monkey, XM_001097616, XM_001097824; Chimpanzee, XM_003309810, XM_526202; Mouse, NM_009895; Rat, NM_031804; Cattle, NM_001046586; Chicken, NM_204626; Western clawedfrog, NM_00113689; Zebrafish, NM_001076617; Rainbow, NM_001171852.
The 3,476 bp, 806 bp, and 1,004 bp fragments were obtained from genomic DNA using primer C-12, C-23, and C-33, respectively. Then, these three sequences and the 1,913 bp cDNA sequence were assembled into a 5,409 bp genomic sequence of pCISH gene which was consisted of three exons and two introns. Further analysis found that the ATG start codon was located in exon1 and the exon 3 contains the TGA stop codon. In addition, by alignment of this 5,409 bp fragment with the porcine genomic sequence (Sscrofa10.2) using BLAT (http://www.ensembl.org), the pCISH gene was assigned to porcine chromosome 13.
Intracellular distribution of the pCISH
The cellular location of pCISH was determined by fluorescence and confocal analysis of PK15 cells transiently transfected with pEGFP-C1-CISH. After labeling nuclei by staining with Hoechst 33342, CISH-EGFP fusion proteins were found in distribution throughout the cytoplasm (Figure 2). This result was consistent with the previously information of primarily cytoplasmic localization of rat CISH in HEK 293t (ATCC) cells [17].
Genetic variation analysis of pCISH
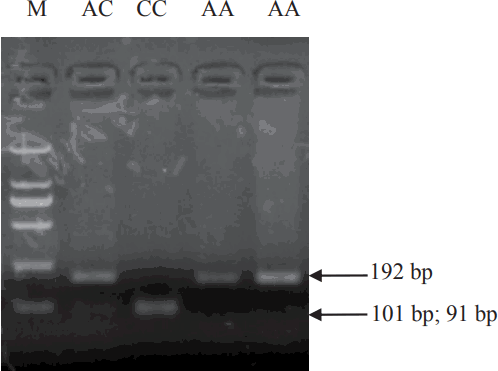
Comparative sequence analysis revealed more than 20 polymorphisms among the pCISH genomic sequence in three pig breeds. SNP A-1575G in promoter region and SNP A2497C in Intron1 could be detected by TaqI and HaeIII PCR-RFLP, respectively, and these SNPs were selected for further association analysis. For the TaqI PCR-RFLP assay, the 1,554 bp PCR product was digested into one fragments of the A allele (1,554 bp) and two fragments of the G allele (1,042 and 512 bp) (Figure 3). For the HaeIII PCR-RFLP, the 192 bp PCR product was digested into one fragments of the A allele (192 bp) and two fragments of the C allele (101 and 91 bp) (Figure 4).
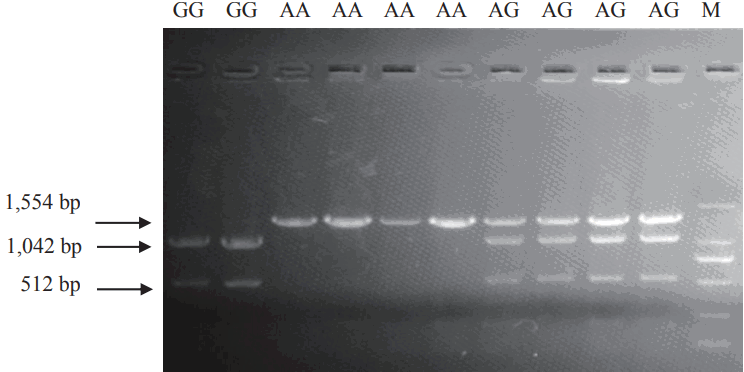
The three different TaqI PCR-RFLP genotypes of pCISH gene. The genotypes (AA, AG, GG) are shown at the top. M: DNA molecular marker DL2000. PCR-RFLP, polymerase chain reaction–restriction fragment length polymorphism; pCISH, porcine cytokine inducible SH2-containing protein.
Statistical analysis
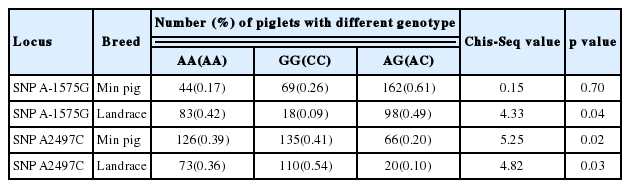
Gen Mod procedure analysis showed that SNP A-1575G was significantly associated with diarrhea index of Min piglets (p< 0.05) (Table 2), while SNP A2497C was significantly associated with the diarrhea trait of both Min pig and Landrace piglets (p< 0.05) (Table 2).
DISCUSSION
Our present study provides the basic structure of pCISH. Comparison with the gene structure of human and rat CISH reveals remarkable similarity among these three species, and they both contained 3 Exons and 2 Introns. The inferred amino acid sequence of porcine CISH showed 93% and 91% identity with that of human and rat respectively. Further analysis showed that pCISH contained a SH2 domain (80–169AA) and a SOCS domain (210–251AA). The SH2 domain and C-terminal SOCS box are characteristic of the SOCS protein family. In the negative regulation of several cytokine pathways, SOCS proteins bind to tyrosine-phosphorylated residues of target proteins via their SH2 domains, then inhibit JAK activity through their N-terminal domains, and are thought to induce the degradation of bound molecules through a conserved SOCS-box motif that interacts with the proteasome [18]. Hence, pCISH might play a role in the cytokine signal transduction of pigs. In the phylogenetic tree of CISH, pig is closer to cattle than other mammals in an evolutionary relationship. As far as we know, research on cattle CISH is extremely scarce, so this present study indicates that it might be interesting to determine the importance of CISH gene in disease resistance of cattle.
In humans, many polymorphisms in CISH gene were observed and some were strongly associated with the susceptibility to diseases caused by diverse infectious pathogens [12–14]. In this study, firstly, we obtained the genomic sequence of pCISH, but observed only two SNPs which could be detected by PCR-RFLP. Statistical analysis indicated that SNP A-1575G was significantly associated with Min piglets diarrhea index (p<0.05), while SNP A2497C was significantly associated with the diarrhea trait of both Min pig and Landrace piglets (p<0.05).
Two important considerations in candidate gene selection are function and position [19]. To assess the biological function of porcine SOCS gene family, Delgado-Ortega et al [20] evaluated the expression pattern of SOCS genes in 10 Selected tissues (kidney, large intestine, liver, lung, mesenteric lymph node, small intestine, spleen, stomach, trachea, and thymus) of twenty two-months-old healthy piglets and found that pCISH gene was highly expressed in large intestine, small intestine, spleen and thymus, suggesting that this gene might play a role in porcine intestine immune responses. In this research, the SNP A-1575G and SNP A2497C resided in the 5′ upstream region and Intron1, respectively. Although the SNP in these regions did not directly alter any amino acid residue, they might play a role in regulating gene expression and thus their constituent SNPs might be directly related to functional variation. For example, a SNP (G3072A) in intron3 of insulin-like growth factor 2 (IGF2) gene was significantly associated with muscle growth in the pig by regulating the expression of IGF2 mRNA in postnatal muscle [21]. So, in further study, we intend to reveal whether these mutations have effects on the transcription of pCISH gene. To account for the effect of position, we compared the position of pCISH gene and the quantitative trait loci (QTL) for anti-disease or immunity traits. The pCISH gene was located within 27 to 28 MB of porcine chromosome 13 (SSC13) by comparing sequence obtained in this study from the database of porcine genome sequence (Sscrofa10). Interestingly, when browsing QTLs in the latest released Pig QTL Database, we found two QTL for enterotoxigenic Escherichia coli (ETEC) susceptibility on SSC13 [22], but the pCISH gene does not reside in the region of these QTLs. So, these SNPs might be potential markers for piglet diarrhea traits, even if they do not alter the amino acid residues or transcription factor binding sites of the pCISH gene, suggesting these mutations affect piglet diarrhea traits indirectly, being in linkage disequilibrium with QTLs or another causative polymorphisms affecting piglet diarrhea traits.
In conclusion, we have shown that pCISH was highly conserved at the level of gene structure. Phylogenetic analysis revealed that based on the protein sequence of CISH, pig and cattle were closer than other mammals in evolutionary relationship. Our data also indicated that pCISH was mainly localized in cytoplasm in the PK-15 cells. Furthermore, association analysis revealed that these two detected SNPs significantly influenced piglet diarrhea, suggesting that pCISH gene might be a novel candidate gene for piglet diarrhea traits. However, our results were based on a limited number and breeds of animals, further studies in more pig populations with a larger sample size or different breeds are needed to confirm these preliminary researches.
ACKNOWLEDGMENTS
This study was supported financially by National Natural Science Foundation of China (31301935), Ph.D. Programs Foundation of Ministry of Education of China (20112325120013).
Notes
CONFLICT OF INTEREST
We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.