Association of DNA Methylation Levels with Tissue-specific Expression of Adipogenic and Lipogenic Genes in Longissimus dorsi Muscle of Korean Cattle
Article information
Abstract
Epigenetic factors, such as DNA methylation status, may regulate adipogenesis and lipogenesis, thus affecting intramuscular fat (IMF) deposition in longissimus dorsi muscle (LM) of beef cattle. In Korean cattle steers, the LM consists mainly of muscle tissue. However, the LM tissue also contains IMF. We compared the gene expression levels between the IMF and muscle portions of the LM after tissue separation. Real-time polymerase chain reaction analysis showed that the mRNA levels of both adipogenic peroxisome proliferator-activated receptor gamma isoform 1 (PPARG1) and lipogenic fatty acid binding protein 4 (FABP4) were higher (p<0.01) in the IMF than in the muscle portion of the LM. We determined DNA methylation levels of regulatory regions of the PPARG1 and FABP4 genes by pyrosequencing of genomic DNA. DNA methylation levels of two of three CpG sites in the PPARG1 gene promoter region were lower (p<0.05) in the IMF than in the muscle portion of the LM. DNA methylation levels of all five CpG sites from the FABP4 gene promoter region were also lower (p<0.001) in the IMF than in the muscle portion. Thus, mRNA levels of both PPARG1 and FABP4 genes were inversely correlated with DNA methylation levels in regulatory regions of CpG sites of the corresponding gene. Our findings suggest that DNA methylation status regulates tissue-specific expression of adipogenic and lipogenic genes in the IMF and muscle portions of LM tissue in Korean cattle.
INTRODUCTION
Beef production in Korea mainly focuses on the meat quality, particularly the degree of intramuscular fat (IMF) deposition; adipogenesis and lipogenesis are important processes for IMF deposition. Adipogenic/lipogenic transcriptional networks regulate IMF deposition in beef cattle (Moisa et al., 2014). Japanese Wagyu cattle are well known for their extremely high marbling (Duarte et al., 2013) indicated that intramuscular adipogenesis was enhanced in Wagyu compared with Angus muscle. Adipogenic peroxisome proliferator activated receptor gamma (PPARG), which is a nuclear receptor and transcription factor, regulates lipid metabolism by controlling the expression of various lipogenic genes, such as adipocyte fatty acid binding protein 4 (FABP4), lipoprotein lipase, acyl-CoA synthase, and fatty acid transport protein (Sarjeant and Stephens, 2012). Our recent studies showed that PPARG and FABP4 gene expression is associated with IMF deposition in Korean cattle (Jeong et al., 2013; Ahn et al., 2014).
DNA methylation is an important epigenetic marker of the transcriptionally repressed state of the genes (Jones and Takai, 2001). Epigenetic regulatory mechanisms including DNA methylation are reportedly involved in the transcriptional activation of PPARG during adipogenesis (Musri et al., 2007). Studies have demonstrated the contribution of PPARG promoter DNA methylation to its expression in adipocyte cell culture systems (Noer et al., 2006; Fujiki et al., 2009). DNA methylation status may be one of mechanisms regulating adipogenic/lipogenic gene expression during IMF deposition. However, involvement of DNA methylation in regulation of adipogenesis and lipogenesis in cattle is unknown. DNA methylation status may be one of mechanisms regulating adipogenic/lipogenic gene expression during IMF deposition. In this study, association of DNA methylation levels and PPARG1 and FABP4 gene expression levels were examined in the IMF and muscle portions of the longissimus dorsi muscle (LM) tissues in Korean cattle steers.
MATERIALS AND METHODS
All experimental procedures involving animals were approved by the Chonnam National University Institutional Animal Use and Care Committee (CNUIAUCC), Republic of Korea. The experiments were conducted in accordance with the Animal Experimental Guidelines provided by CNUIAUCC.
Animals and tissue samples
In this study, we used steer LM tissue samples from previous work (Bong et al., 2012). Slaughter age was 846±30 days, and carcass weight was 398±10 kg. We separated the muscle and IMF portion from the intact LM tissues to determine tissue-specific DNA methylation pattern, as previously described (Bong et al., 2012).
RNA extraction and quantitative real-time polymerase chain reaction
To detect expression levels of PPARG1 and FABP4 genes, total RNA was isolated as previously described (Jeong et al., 2013) using TRIzol reagent (Molecular Research Center, Cincinnati, OH, USA). Total RNA was quantified by absorbance at 260 nm, and the integrity of total RNA was verified through agarose gel electrophoresis and ethidium bromide staining of the 28S and 18S bands. Total RNA (0.5 μg) was reverse-transcribed into cDNA using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA).
Real-time polymerase chain reaction (PCR) was performed as previously described (Jeong et al., 2013) using the QuantiTect SYBR Green RT-PCR Master Mix (Qiagen, Valencia, CA, USA) and an Opticon Sequence Detection system (MJ Research, Waltham, MA, USA) with gene-specific primers (Table 1). The ΔΔCT method was used to determine the fold change in mRNA expression relative to the housekeeping gene, ribosomal protein, large, P0.
DNA methylation determination by pyrosequencing of bisulfite-treated genomic DNA
Genomic DNA extraction and analysis of DNA methylation were performed by the DisGene Company (Daejon, Republic of Korea). Briefly, tissue genomic DNA was purified using a NucleoSpin Tissue column (Macherey-Nagel GmbH & Co., Duren, Germany).
To determine DNA methylation levels, target regions of PPARG1 (Figure 1a) and FABP4 (Figure 2a) genes were selected from the CpG islands, which were searched using CpG Island Searcher (USC Norris Comprehensive Cancer Center, USA; http://www.uscnorris.com/cpgislands2/cpg.aspx). Transcription factor binding sites (Figures 1a and 2a) were determined using TFSEARCH (Computational Biology Research Center, National Institute of Advanced Industrial Science and Technology, Japan; http://www.cbrc.jp/research/db/TFSEARCH.html). We tried to find promoter regions of the genes in which important transcription factor binding sites were located that may regulate gene expression.
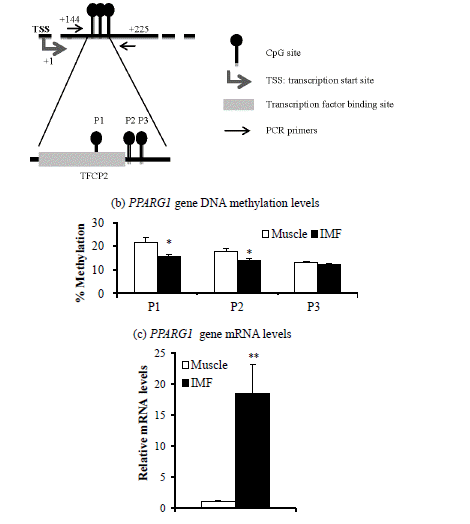
Target area (a), DNA methylation levels (b), and mRNA levels (c) of the peroxisome proliferator activated receptor gamma 1 (PPARG1) gene in intramuscular fat (IMF) and muscle portion of Korean cattle steer longissimus dorsi muscle tissue. (a) DNA methylation assay target area of the PPARG1 gene promoter region and transcription factor binding sites. TFCP2, transcription factor CP2. (b) DNA methylation levels were determined by pyrosequencing of bisulfite-treated DNA. Values are the mean+SE (n = 5). (c) The mRNA levels were determined by real-time PCR and normalized against a housekeeping gene. Muscle portion data were normalized to 1.0. Values are the mean+SE (n = 10). * p<0.05; ** p<0.01. PCR, polymerase chain reaction; SE, standard errors.
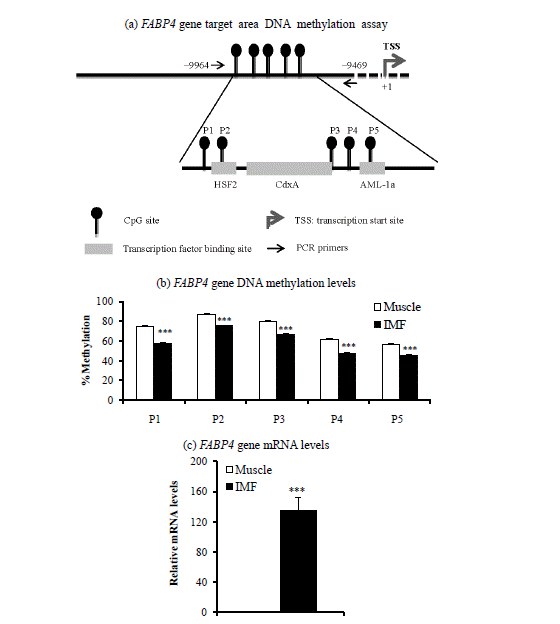
Target area (a), DNA methylation levels (b), and mRNA levels (c) of the fatty acid binding protein 4 (FABP4) gene in intramuscular fat (IMF) and the muscle portion of Korean cattle steer longissimus dorsi muscle tissue. (a) DNA methylation assay target area in the FABP4 gene promoter region and transcription factor binding sites. HSF2, heat shock factor 2; CdxA, caudal-related homeobox A; AML-1a, acute myeloid leukemia-1a. (b) DNA methylation levels were determined by pyrosequencing bisulfite-treated DNA. Values are the mean+SE (n = 5). (c) The mRNA levels were determined by real-time PCR and normalized with a housekeeping gene. Muscle portion data were normalized to 1.0. Values are the mean+SE (n = 10). *** p<0.001. PCR, polymerase chain reaction; SE, standard errors.
Bisulfite treatment of genomic DNA was performed using the EpiTect Bisulfite Kit (Qiagen, USA). The bisulfite-treated DNA was amplified by PCR using primers indicated in Table 2. Pyrosequencing of PCR products was done using primers shown in Table 2. The degree of methylation at each CpG site was determined from the ratio of thymine (T) and cytosine (C) by the following equation:
Statistical analysis
Data are expressed as means±standard errors. Differences between muscle and IMF portions of the LM tissues were analyzed by the general linear model procedure in SAS (SAS Inst. Inc., Cary, NC, USA), and model included tissue type and a random error.
RESULTS AND DISCUSSION
Real-time PCR analysis showed that mRNA levels of both PPARG1 (Figure 1b; p<0.01) and FABP4 (Figure 2b; p<0.001) genes were higher in the IMF portion than in the muscle portion of the LM. Our recent study also showed higher PPARG1 (Jeong et al., 2013) and FABP4 (Ahn et al., 2014) mRNA levels in IMF than in the muscle portion of the LM.
Next, we determined DNA methylation levels within CpG island promoter regions of the PPARG1 and FABP4 genes. DNA methylation levels of two of three CpG sites from the PPARG1 gene regulatory region (+144 to +225) were lower (p<0.05) in the IMF portion than in the muscle portion of the LM (Figure 1c). DNA methylation levels of all five CpG sites from promoter regions (−9,664 to −9,469) of the FABP4 gene were lower (p<0.001) in the IMF portion than in the muscle portion (Figure 2c). Thus, transcription levels of both PPARG1 and FABP4 genes were inversely correlated with DNA methylation levels of regulatory regions of CpG sites of the corresponding gene.
Two types of PPARG splice variants, PPARG1 and PPARG2, have been identified in several species, including mouse (Zhu et al., 1993), human (Elbrecht et al., 1996), and bovine species (Sundvold et al., 1997). Differential expression of two types of PPARG has been reported in several studies: PPARG1 mRNA was expressed to a higher levels compared to PPARG2 mRNA in human adipose tissues (Vidal-Puig et al., 1997; Yanase et al., 1997). We found that PPARG1 mRNA levels in the IMF portions were 18-fold higher than muscle portion of the LM, whereas PPARG2 mRNA levels in the IMF were only 2 fold higher than muscle portion (unpublished data), although similar mRNA levels of both transcripts were detected in bovine fat tissues in other study (Sundvold et al., 1997). Thus, we have chosen PPARG1 rather than PPARG2 for DNA methylation analysis. In a previous study, the PPARG2 gene promoter in 3T3-L1 preadipocytes was hypermethylated, but was also progressively demethylated upon the induction of differentiation, which was accompanied by an increase in mRNA expression (Fujiki et al., 2009). They showed that PPARG gene expression was inhibited by methylation of its promoter region. The CpGs within the FABP4 promoter were methylated in muscle progenitor cells, whereas CpGs were relatively unmethylated in adipose stem cells (Sorensen et al., 2010). A recent study also showed that treatment with 5-aza-29-deoxycytidine, a demethylating agent, decreased adipocyte differentiation, resulting in the downregulation of PPARG2 and FABP4 gene expression (Zych et al., 2013).
Methylation at specific CpG positions could influence the affinity for specific transcription factors toward DNA molecules (Deaton and Bird 2011). We found that DNA methylation levels of the first two PPARG1 promoter CpG sites (+187, +203) were lower in the IMF portion than those in the muscle portion of steers. These regions (+144 to +225) are located on the first exon of PPARG1 and contains a CP2 transcription factor binding site (TFCP2). Decreased DNA methylation on specific CpG sites could permit the induction of PPARG1 gene transcription. Thus, DNA methylation status may alter TFCP2 binding activity at CpG sites, regulating PPARG1 gene transcription. Association of DNA methylation status with transcriptional control of the PPARG1 gene via the TFCP2 has not been reported.
We also found that the IMF portion had lower DNA methylation levels in all five CpG sites of the FABP4 gene upstream region (−9,664 to −9469) compared to the muscle portion, whereas the FABP4 gene mRNA level was higher in IMF than in the muscle portion of intact LM in the current study as well as in our recent study (Ahn et al., 2014). The upstream region of the FABP4 gene in which we measured DNA methylation status contains transcription factor binding sites for several transcription factors: heat shock transcription factor 2 (HSF2) on CpG site 2, caudal-related homeobox A (CdxA) on CpG site 3, and acute myeloid leukemia-1a (AML-1a) on CpG site 5. Thus, DNA methylation status may alter transcription factor binding activities at these CpG sites, regulating transcription of the FABP4 gene. Whether DNA methylation status alters binding activity of these transcription factors on the FABP4 gene promoter is unknown.
In this study, the differences of DNA methylation levels between IMF and muscle portion were about 10%. In contrast, the differences of PPARG1 and FABP4 gene expression levels between IMF and muscle portion were about 10 to 100 times (Figure 1c and Figure 2c). Our study suggests that minor difference of DNA methylation status of PPARG2 and FABP4 promoter may profoundly affect gene expression levels. Similarly, 5′-aza-cytideine, an inhibitor of DNA methylation, increased PPARG2 mRNA levels over 20 times, although methylation levels were decreased about 2 times from 40% to 20% (Fujiki et al., 2009). They suggest that promoter demethylation is not the only factor controlling PPARG2 expression.
In conclusion, DNA methylation status may regulate tissue-specific differential expression of PPARG1 and FABP4 genes in the IMF and muscle portion of LM tissues.
ACKNOWLEDGMENTS
This study was supported by a grant from the Next Generation BioGreen 21 Program (No. PJ00819103), Rural Development Administration, Republic of Korea.

