Degradation of Phytate Pentamagnesium Salt by Bacillus sp. T4 Phytase as a Potential Eco-friendly Feed Additive
Article information
Abstract
A bacterial isolate derived from soil samples near a cattle farm was found to display extracellular phytase activity. Based on 16S rRNA sequence analysis, the strain was named Bacillus sp. T4. The optimum temperature for the phytase activity toward magnesium phytate (Mg-InsP6) was 40°C without 5 mM Ca2+ and 50°C with 5 mM Ca2+. T4 phytase had a characteristic bi-hump two pH optima of 6.0 to 6.5 and 7.4 for Mg-InsP6. The enzyme showed higher specificity for Mg-InsP6 than sodium phytate (Na-InsP6). Its activity was fairly inhibited by EDTA, Cu2+, Mn2+, Co2+, Ba2+ and Zn2+. T4 phytase may have great potential for use as an eco-friendly feed additive to enhance the nutritive quality of phytate and reduce phosphorus pollution.
INTRODUCTION
Phytate or phytic acid (myo-inositol hexakisphosphate; InsP6), the primary phosphorus reservoir of higher edible plants such as cereals, legumes and oil seeds (Haefner et al., 2005) is little or poorly metabolized in monogastric animals including swine, poultry and humans because they have insufficient phytate-degrading activity in their digestive tracts (Shobirin et al., 2007). Consequently, the non-utilized phytate phosphorus from animal manures contributes to environmental pollution (Luo et al., 2007; Kebreab et al., 2011). Moreover, phytate is an anti-nutritive agent that forms insoluble complexes with divalent metal ions such as Fe2+, Ca2+, Mg2+, and Zn2+, and binds to positively charged proteins, thereby reducing their bioavailability (Oh et al., 2004; Fu et al., 2008; Gibson et al., 2010).
Phytases are present in plants, certain animal tissues, and microorganisms (Kim et al., 1998). They are a unique class of phosphatases that catalyze the stepwise hydrolysis of phytate to lower inositol phosphate esters and inorganic phosphate (Oh et al., 2004). Phytase application can lessen the amount of inorganic phosphate that is presently excessively added to animal diets, improve mineral availability, and reduce phosphorus pollution (Haefner et al., 2005; Adeola and Cowieson, 2011). Virtually, phytases are now added routinely to swine and poultry diets as an economical aid in the digestibility of phosphorus and an important measure for environmental protection (Selle and Ravindran, 2007). Currently, a number of microbial phytases with different enzymatic properties are commercially available (Sulabo et al., 2011). Catalytically, most bacterial, fungal, and plant phytases belong to the family of histidine acid phosphatases (HAP) typically characterized by a conserved active site motif, RHGXRXP (Lei and Porres, 2003). Meanwhile, a new subgroup of phytases, β-propeller phytase that does not contain the conserved active site motif and exhibits unusual Ca2+-dependent catalytic property with strict substrate specificity has been discovered from Bacillus strains (Mullaney and Ullah, 2003; Fu et al., 2008). To date, research aimed at characterizing Bacillus phytase has been solely conducted with the dodecasodium salt of phytate (Na-InsP6) as a main substrate (Powar and Jagannathan, 1982; Kerovuo et al., 1998; Kim et al., 1998; Choi et al., 2001; Gulati et al., 2007), which is not a genuine substrate in biological systems (Oh et al., 2004) and which is appropriate solely for screening HAP phytases capable of hydrolyzing the metal-free form of phytate at acidic pH (Oh et al., 2001).
The current study is the first report on the general catalytic properties of Bacillus sp. T4 phytase acting on pentamagnesium salt of phytate (magnesium phytate, Mg-InsP6), which was recently identified as the most physiologically relevant substrate within the cytosolic and nuclear compartments of eukaryotic cells (Torres et al., 2005; Cho et al., 2006).
MATERIALS AND METHODS
Chemicals, organism and screening
Na-InsP6 and Aspergillus ficuum phytase were purchased from Sigma-Aldrich (St. Louis, MO, USA). Mg-InsP6 was prepared as previously described (Torres et al., 2005). Unless otherwise stated, other analytical chemicals used in this study were also purchased from Sigma-Aldrich. The bacterial isolate, T4 derived from soil samples near a cattle farm was provided by the Department of Agricultural Biotechnology, Seoul National University (South Korea). Screening for phytase activity was done on the selected differential agar (2% (w/v) glucose, 0.5% (w/v) (NH4)2SO4, 0.07% (w/v) KCl, 0.01% (w/v) MgSO47H2O, 0.01% (w/v) NaCl, 0.01% (w/v) CaCl22H2O, 0.001% (w/v) MnSO44H2O, 0.001% (w/v) FeSO47H2O, 0.5% (w/v) Na-InsP6, and 1.5% (w/v) bacto agar (Difco, Detroit, MI, USA); (pH 6.5)] at 30°C on the basis of a previously described two-step counterstaining method using cobalt chloride and ammonium molybdate/ammonium vanadate solution (Bae et al., 1999).
Taxonomic identification of strain T4
Genomic DNA was extracted from strain T4 using the FastDNA kit (Qbiogene, Montreal, PQ, Canada) according to the manufacturer’s protocol. The 16S rRNA gene was amplified from genomic DNA by polymerase chain reaction (PCR) using the universal primers 27F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492R (5’-GGTTACCTTGTTACGACTT-3’) (William et al., 1991). The amplified 1,449 bp sequences were determined by an automated ABI PRISM 3730 XL DNA analyzer (Applied Biosystems, Foster city, CA, USA). The resulting sequences were compared with the GenBank database (NCBI) using BLAST (Altschul et al., 1990). Sequences showing a relevant degree of similarity were imported into the CLUSTAL W program (Thompson et al., 1994) and aligned. The evolutionary distances with other strains of Bacillus were computed using the Maximum Composite Likelihood method (Tamura et al., 2004) and the phylogenetic relationships were determined using the software MEGA version 4.0 (Tamura et al., 2007).
Nucleotide sequence accession numbers
The nucleotide sequence of the 16S rRNA gene has been deposited in the GenBank database under Accession No. JF508860.
Partial purification of the enzyme
For the production of phytase, strain T4 was cultured in 25 ml of the wheat bran extract medium (1% (v/v) wheat bran extract, 1% (w/v) casein hydrolysate, 0.04% (w/v) (NH4)2SO4, 0.05% (w/v) KH2PO4, 0.04% (w/v) K2HPO4, 0.2% (w/v) CaCl22H2O, 0.02% (w/v) MgSO47H2O; (pH 6.5)) prepared by the method of Powar and Jagannathan (1982) in an Erlenmeyer flask of 250 ml capacity for 24 h at 37°C. Then, 1 L of the same medium in two Erlenmeyer flasks of 2 L capacity was aseptically inoculated with 2% seed culture broth and aerobically grown with vigorous shaking (220 rpm) for 96 h at 37°C. The culture medium containing secreted phytase was centrifuged (9,000×g; 30 min; 4°C) to remove cells, and then protein in the supernatant was precipitated with ammonium sulfate (80% saturation) in presence of 2 mM CaCl2. The pellet was dissolved in 25 mM Tris-HCl (pH 8.0) containing 2 mM CaCl2 and dialyzed overnight against 25 mM Tris-HCl (pH 8.0) at 4°C. The dialyzed solution was used as the phytase source throughout this work to examine its catalytic properties.
Phytase differential staining
The activity staining of T4 phytase was carried out as previously described (Bae et al., 1999) with the following modifications. Briefly, T4 strain was streaked onto phytase screening agar medium (0.5% (w/v) sodium phytate, 2% (w/v) glucose, 0.5% (w/v) (NH4)2SO4, 0.2% (w/v) CaCl2·2H2O, 0.01% (w/v) MgSO4·7H2O, 0.01% (w/v) NaCl, 0.07% (w/v) KCl, 0.001% (w/v) MnSO4·4H2O, 0.001% (w/v) FeSO4·7H2O, 1.5% (w/v) bacto agar, pH 6.5), and incubated at 37°C for 96 h. After growth, the colonies were washed from the agar surface and the plate was flooded with 2% (w/v) cobalt chloride solution for 5 min. Then, the cobalt chloride solution was replaced with a freshly prepared solution containing equal volumes of a 6.25% (w/v) ammonium molybdate solution and a 0.42% (w/v) ammonium vanadate solution. Following 5 min incubation, the ammonium molybdate/ammonium vanadate solution was removed and the plate examined for zones of clearing.
Phytase activity assay and substrate specificity
Unless otherwise stated, phytase assay was performed in 1 ml of 50 mM Bis-Tris buffer (pH 6.0) containing 1 mM Mg-InsP6 at 40°C for 1 h. The released inorganic phosphates were measured by a modified method of Heinonen and Lahti (1981) with a freshly prepared acetone ammonium molybdate (AAM) reagent consisting of acetone, 5N sulfuric acid, and 10 mM ammonium molybdate (2:1:1, v/v). Two milliliters of the AAM solution and, thereafter, 0.2 ml of 1 M citric acid were added to the phytase assay mixture. Absorbance was read at 355 nm after blanking the spectrophotometer with an appropriate control. The enzyme activity with other phosphorylated compounds such as adenosine 5’-triphosphate (ATP), adenosine 5’-monophosphate (AMP), ribose-5-phosphate, glucose-6-phosphate and 2,3-bisphosphoglycerate (2,3-BPG) was measured under the standard assay condition using 1 mM of each substrate. Acid phosphatase activity was determined using p-nitrophenyl phosphate as previously described (Greiner et al., 1993). The release of p-nitrophenol was measured by the absorbance of 405 nm. One unit (U) of enzyme activity was defined as the amount of enzyme to catalyze the release of 1 μmol of inorganic phosphate or p-nitrophenol per minute under the defined assay conditions.
Effect of pH on enzyme activity
The profile of pH versus activity was investigated by measuring the phytase activity at 30°C and pH range of 3.0 to 8.5 using the following buffers such as 50 mM glycine-HCl (pH 3), 50 mM sodium acetate (pH 4 to 5.5), 50 mM Bis-Tris-HCl (pH 6 to 7) and 50 mM Tris-HCl (pH 7.4 to 8.5) for 1 h.
Effect of temperature on enzyme activity and thermal stability
The profile of temperature versus activity was examined at the optimum pH in a temperature range of 0 to 80°C. The thermal stability was measured by assessing the residual activity after pre-incubation of the enzyme in 50 mM Bis-Tris-HCl (pH 6.0) for 30 min at various temperatures ranging from 30 to 80°C in absence or presence of 5 mM CaCl2.
Effect of metal ions and chemicals on enzyme activity
The effects of various metal ions and chemicals on phytase activity were determined at 40°C for 1 h in the presence of 1 mM of Ba2+, Mg2+, Ca2+, Co2+, Zn2+, Ni2+, Cu2+, Mn2+, ethylenediaminetetraacetic acid (EDTA), sodium dodecyl sulfate (SDS) or phenylmethylsulfonyl fluoride (PMSF).
RESULTS AND DISCUSSION
Identification of isolated strain T4
To identify the bacterial isolate T4 exhibiting phytase activity (Figure 1), we cloned the 16S rRNA gene and compared the sequence with those available in the database. A phylogenic tree based on the 16S rRNA gene sequences from 10 Bacillus strains showed that the isolate T4 shared 99.5% sequence identity with the type strain, Bacillus subtilis DSM 10T (Figure 2). Thus, it was named Bacillus sp. T4.

Qualitative plate activity staining of the phytase produced by Bacillus sp. T4. The clearing zones marked as white arrows indicate the phytase activity.
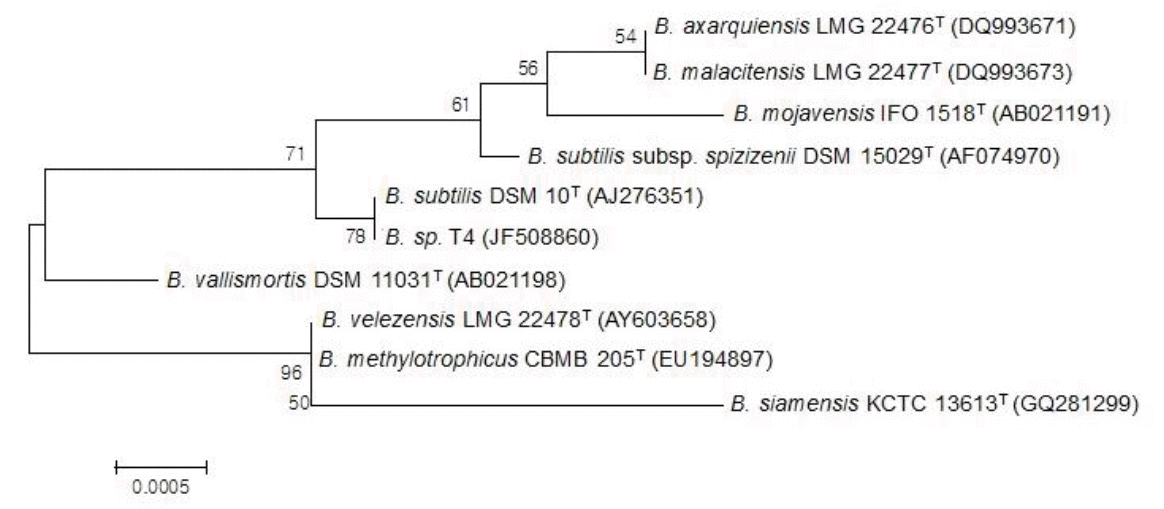
Phylogenic relationship of the 16S rRNA sequences of Bacillus sp. T4 with other type strains of Bacillus. Bootstrap values (based on 1,000 trials and only values >50%) are shown at the nodes. The GenBank accession numbers are indicated in parentheses. Bar, 0.0005 substitutions per nucleotide position. B. represents the abbreviation of Bacillus.
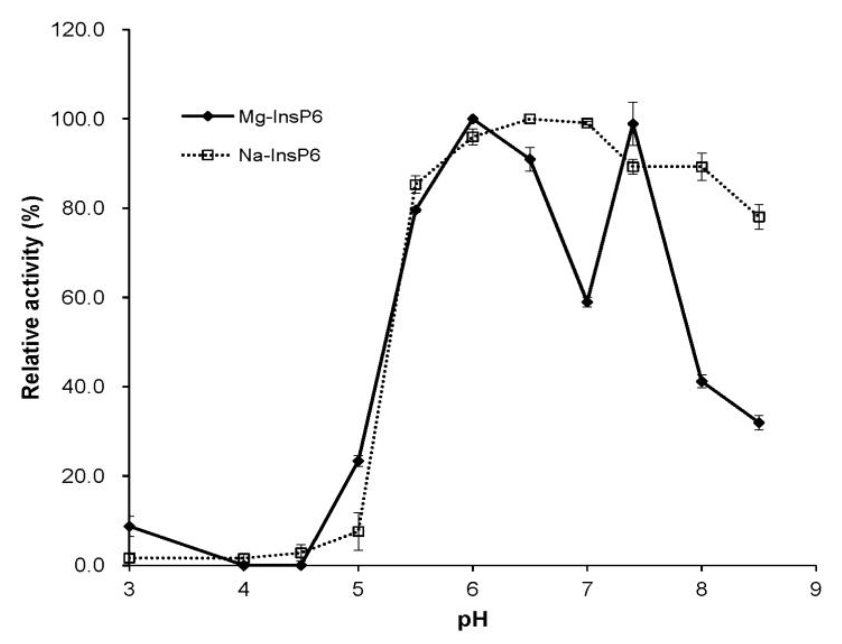
Effect of pH on enzyme activity
The optimal activity of T4 phytase toward Na-InsP6 was found at the pH range of 6.5 to 8.0 (Figure 3), as previously noted in many Bacillus strains (Oh et al., 2004). However, T4 phytase had a characteristic bi-hump two pH optima of 6.0 to 6.5 and 7.4 for Mg-InsP6 (Figure 3). This was due to the dip in activity at pH 7.0, as previously observed in avian phytase (also known as multiple inositol polyphosphate phosphatase) (Cho et al., 2006). The occurrence of phytases showing more than one pH optima peak has been reported (Tomschy et al., 2000; Casey and Walsh, 2003). Their unusual pH activity profiles have been explained as possible buffer effect or an artifact (Mullaney et al., 2002).
Effect of temperature on enzyme activity and thermal stability
As shown in Figure 4, without Ca2+, the optimum temperature of T4 phytase was 40°C, which was consistent with that of the Bacillus sp. KHU-10 phytase (Choi et al., 2001) and was very close to the gastrointestinal temperature of pig or poultry (37 to 40°C) for application of the enzyme as a feed supplement (Lei and Porres, 2003). Thermostable phytases from Bacillus laevolacticus and Bacillus sp. DS11 exhibit maximal activities at 70°C, but 50% and 60% of activity was lost at 40°C, respectively (Kim et al., 1998; Gulati et al., 2007). In the presence of Ca2+, optimal activity for T4 phytase occurred at 50°C (Figure 4), which was somewhat different from the previous description of optimal activity of Bacillus sp. KHU-10 phytase at 60°C (Choi et al., 2001). As for thermal stability, T4 phytase was relatively stable up to 40°C, but 90% of the initial activity was lost at 50°C in the absence of Ca2+, whereas 70% of the activity still remained at 50°C in the presence of Ca2+ (Figure 5). Previous results indicated that Ca2+ acts as an essential activator for Bacillus phytases and has an important effect on thermal stability of the enzymes (Oh et al., 2001; Fu et al., 2008). Similarly, a new alkaline β-propeller phytase (PhyA115) recently characterized from the insect symbiotic bacterium, Janthinobacterium sp. TN115, was unstable without Ca2+, losing about 90% of the activity after incubation at 55°C for 4 min, but in the presence of Ca2+, the enzyme retained more than 70% of the activity at 55°C for 30 min (Zhang e al., 2011).
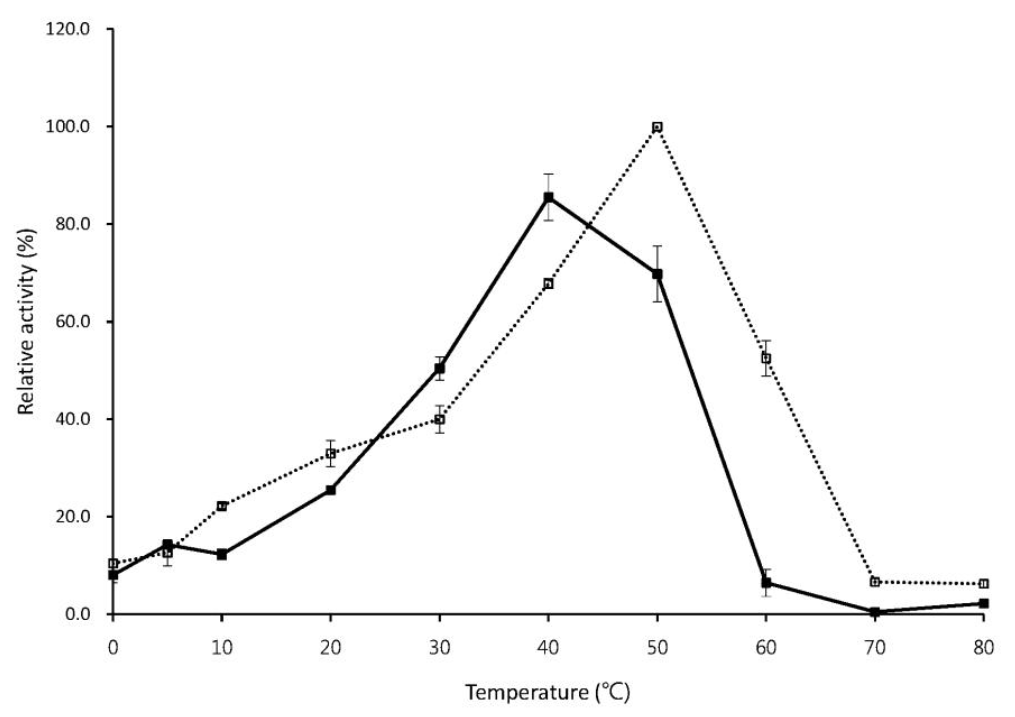
Temperature activity profiles of T4 phytase. Symbols represent the phytase activity in absence of 5 mM CaCl2 (closed squares with solid line) and the phytase activity in presence of (open squares with dotted line). Relative activity is expressed as a percentage of the maximum (100% of relative activity in the phytase in presence of 5 mM CaCl2 equates to 0.047 U/ml). Data represent means and standard errors from three experiments.

Thermal stability of T4 phytase. Symbols represent the residual phytase activity in absence of 5 mM CaCl2 (closed squares with solid line) and the phytase activity in presence of (open squares with dotted line). Residual activity is expressed as a percentage of the maximum (100% of residual activity in the phytase in absence of 5 mM CaCl2 and in presence of equates to 0.035 and 0.026 U/ml, respectively). Data represent means and standard errors from three experiments.
Substrate specificity
As shown in Figure 6, T4 phytase displayed high selectivity for Mg-InsP6 with 1.67-fold higher activity in comparison to Na-InsP6. The enzyme also had hydrolytic activity on some phosphate conjugates such as ATP, AMP and p-nitrophenyl phosphate, which was in good agreement with the phytase from Bacillus laevolacticus (Gulati et al., 2007). In contrast, the phytases from Bacillus subtilis (Powar and Jagannathan, 1982), Bacillus DS11 (Kim et al., 1998) and Bacillus KHU-10 (Choi et al., 2001) exhibited absolutely strict substrate specificity for the phytate substrate. It was interesting to note that T4 phytase showed no phophatase activity on the substrate, 2,3-bisphosphoglycerate (2,3-BPG), a main allosteric effector of hemoglobin (Figure 6), because avian multiple inositol polyphosphate phosphatase classified into the HAP phytase family instead hydrolyzes 2,3-BPG at a rate 1.6-fold higher than the phytate substrate (Cho et al., 2008). On the other hand, in our study, the activity (0.065 U/ml at pH 6.0 and 40°C) of another HAP phytase, fungal Aspergillus ficuum phytase, toward 2,3-BPG was higher than that (0.043 U/ml) of phytate, suggesting that the HAP phytases may share the trait to hydrolyze 2,3-BPG, unlike β-propeller phytases from Bacillus strains.
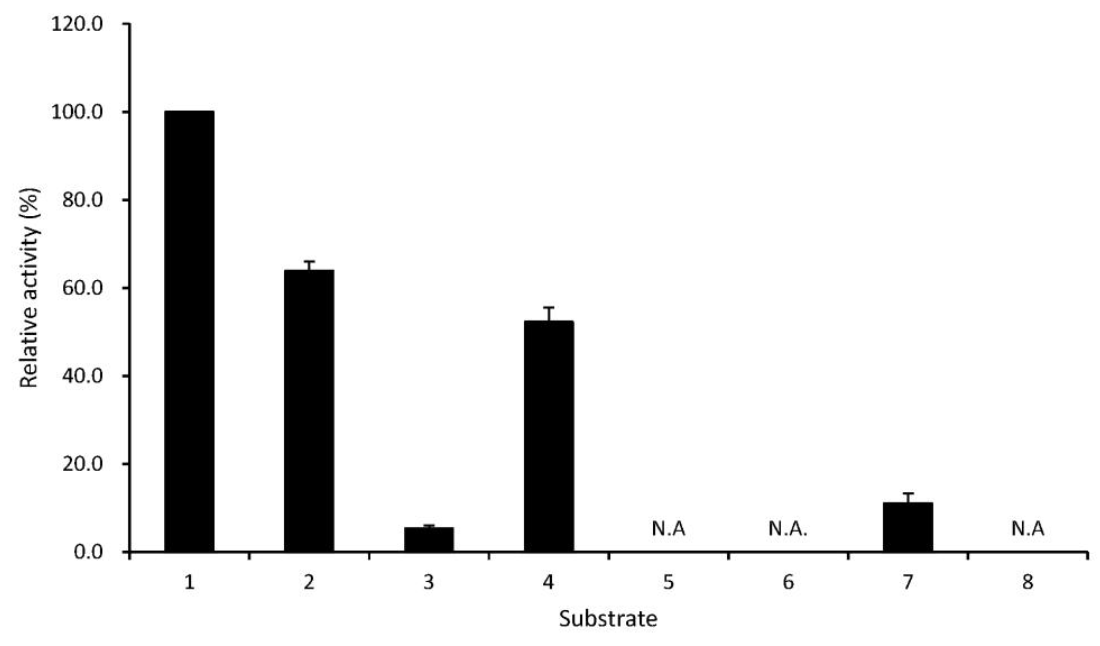
Substrate specificity of T4 phytase. Relative activity is expressed as a percentage of the maximum (100% of relative activity in the phytase equates to 0.037 U/ml). Data represent means and standard errors from three experiments. NA = No activity. 1 = Mg-InsP6; 2 = Na-InsP6; 3 = ATP; 4 = AMP; 5 = ribose-5-phosphate; 6 = glucose-6-phosphate; 7 = p-nitrophenyl phosphate; 8 = 2,3-bisphosphoglycerate.
Effect of metal ions and chemicals on enzyme activity
As shown in Figure 7, among the tested metal ions, T4 phytase activity was fairly reduced (40% to 65% of the control activity) in the presence of Cu2+, Mn2+, Co2+, Ba2+ and Zn2+, which was similar to previous result from Bacillus sp. KHU-10 phytase, except for Zn2+ (Choi et al., 2001). Ca2+ showed a slight stimulatory effect. Instead, no important effect on the T4 phytase activity was observed with Mg2+ and Ni2+. The enzyme was strongly inhibited by EDTA (Figure 7), as previously described (Kerovuo et al., 2000), suggesting that it may be a metalloenzyme. Indeed, Bacillus phytases are catalytically active only when they are structurally associated with Ca2+ (Oh et al., 2001; Tran et al., 2011; Zeng et al., 2011). On the other hand, the activity of Aspergillus niger van Teighem phytase is enhanced up to about 50% by EDTA (Vats and Banerjee, 2005). The enzyme activity was not greatly affected by SDS and PMSF (Figure 7), unlike Aspergillus niger van Teighem phytase, which was sensitive to SDS and PMSF, resulting in the loss of 94% and 68%, respectively (Vats and Banerjee, 2005).
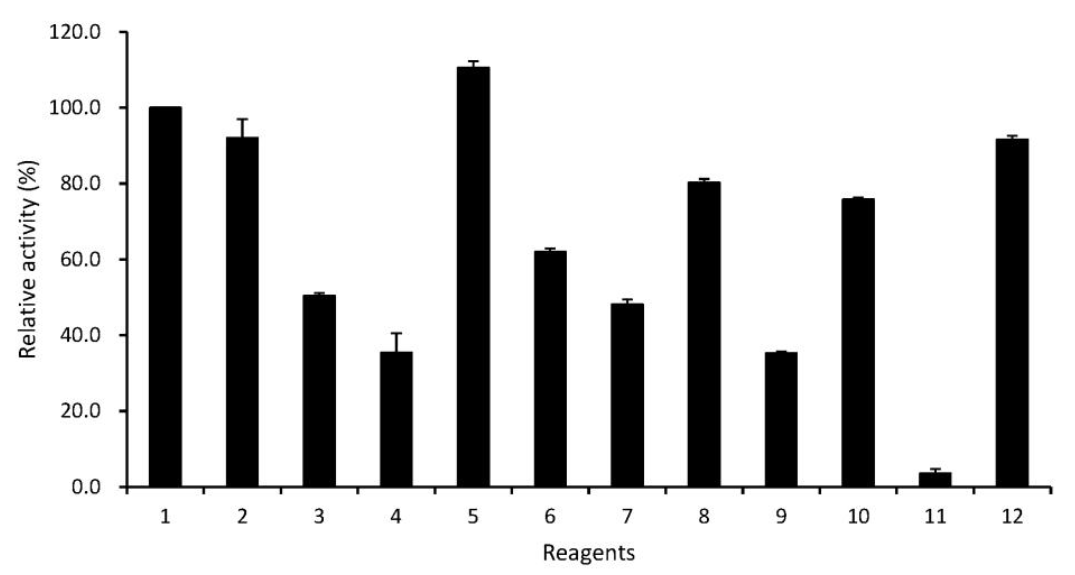
Effect of metal ions and chemicals on T4 phytase activity. Relative activity is expressed as a percentage of the maximum (100% of relative activity in the phytase equates to 0.038 U/ml). Data represent means and standard errors from three experiments. 1 = Control (no addition) ; 2 = Mg2+; 3 = Cu2+; 4 = Mn2+; 5 = Ca2+; 6 = Co2+; 7 = Ba2+; 8 = Ni2+; 9 = Zn2+; 10 = SDS; 11 = EDTA; 12 = PMSF.
Today, the feed industry mainly ascribes the rapidly expanding global market for animal feed enzymes to increased use of exogenous phytases (Sulabo et al., 2011). In conclusion, the T4 phytase may have great potential for use as an eco-friendly feed additive to enhance the nutritive quality of phytate and reduce phosphorus pollution, because it possesses high selectivity on the genuine phytate substrate, Mg-InsP6, and desirable activity profiles at physiologically-relevant pH and temperature. More detailed characterization of the enzyme such as gene cloning, protein engineering, and fermentation technology is warranted to maximize the catalytic efficiency and productive yield of the enzyme.
ACKNOWLEDGEMENT
This paper was supported by Konkuk University in 2012.