Molecular Characterization and Expression Analysis of S6K1 in Cashmere Goats (Capra hircus)
Article information
Abstract
p70 ribosomal S6 kinase (p70S6K) can integrate nutrient and growth factor signals to promote cell growth and survival. We report our molecular characterization of the complementary DNA (cDNA) that encodes the goat p70S6K gene 40S ribosomal S6 kinase 1 (S6K1) (GenBank accession GU144017) and its 3′ noncoding sequence in Inner Mongolia Cashmere goats (Capra hircus). Goat S6K1 cDNA was 2,272 bp and include an open reading frame (ORF) of 1,578 bp, corresponding to a polypeptide of 525 amino acids, and a 694-residue 3′ noncoding sequence with a polyadenylation signal at nucleotides 2,218 to 2,223. The relative abundance of S6K1 mRNA was measured by real-time PCR in 6 tissues, and p70S6K expression was examined by immunohistochemistry in heart and testis. The phosphorylation of p70S6K is regulated by mitogen-activated protein kinase (MAPK) signaling in fetal fibroblasts.
INTRODUCTION
p70 ribosomal protein S6 kinase 1 (S6K1, p70S6K) is a member of the AGC (cyclic AMP-dependent protein kinase, cyclic GMP-dependent protein kinase, and protein kinase C) subfamily of serine/threonine protein kinases. p70S6K is regulated in response to cytokines, nutrients, and growth factors.
p70S6K is involved in protein synthesis, cell growth and apoptosis, embryonic development, and cancer (Fenton and Gout, 2011; Brian et al., 2012; Laplante and Sabatini, 2012). Recent studies have examined p70S6K with regards to cell differentiation (Carnevalli et al., 2010; Hamilton et al., 2010), metabolism (Castañeda et al., 2012; Tandon et al., 2011), autophagy (Wang et al., 2012; Xu et al., 2012), diabetes (Melnik, 2012; Moreno-Navarrete et al., 2012), obesity (Medeiros et al., 2011; Melnik, 2012), aging (Selman et al., 2009; Zoncu et al., 2011), and various diseases and disorders. p70S6K also regulates cellular and organismal physiology.
Due to alternative splicing and differential use of translational start sites, the human S6K1 gene encodes two isoforms, p70 and p85. p85 is longer than p70 by 23 amino acids (Grove et al., 1991) that include a nuclear localization sequence (NLS) in the amino terminal region. Thus, p70 localizes primarily to the cytoplasm, whereas p85 is nuclear (Coffer and Woodgett, 1994; Reinhard et al., 1994). The two classes are believed to mediate cell cycle progression, for which p70S6K activity is essential.
The past several years have witnessed significant gains in our understanding of the stepwise activation of p70S6K by multisite phosphorylation. Like other AGC kinase family members, catalytic activation of p70S6K requires dual phosphorylation of a critical residue in the activation loop (T-loop; T229) and a hydrophobic motif (HM; T389) in its catalytic kinase domain. However, the C-terminal autoinhibitory domain (AID) suppresses catalytic activation by blocking upstream kinase-mediated phosphorylation of the T-loop and HM (Ragan et al., 2008).
Activation of p70S6K requires a complex, ordered series of conformational changes and phosphorylation reactions, which are initiated by an calcium-dependent priming step (Hannan et al., 2003) that releases the interaction between the C-terminal autoinhibitory and N-terminal domains. Next, the autoinhibition of the multisite Ser-Thr phosphorylation of the AID is relieved (S411, S418, T421, and S424) (Ragan et al., 2008), and S371 and T389, which lie in a hydrophobic motif, are phosphorylated through a mTOR-dependent pathway (Saitoh et al., 2002). Finally, in the catalytic domain activation loop, T229 is phosphorylated by PDK1 (3-phosphoinositide-dependent protein kinase) (Pullen et al., 1998).Two models about activation of p70S6K have been proposed: a conventional, widely accepted model and an alternate model (Brian et al., 2012).
Many studies have examined the upstream regulators of mTORC1 signaling and crosstalk between mTOR and PI3K and MEK1/2 signaling pathways. The PI3K and MEK1/2 signaling pathways converge through the tuberous sclerosis protein 1/2 complex (TSC1/2) and the small GTPase Rheb on mTOR (Laplante and Sabatini, 2012; Wullschleger et al., 2006). Nevertheless, as one of the downstream effectors of mTORC1, the relationship between MEK/ERK and p70S6K is undefined (Nawroth et al., 2011; Rolfe et al., 2005).
The mitogen-activated protein kinase (MAPK) signaling pathways mediate many processes, such as growth, differentiation, and stress responses. The prototypical MAPK signaling pathway is the Raf/MEK/ERK cascade, which regulates cell growth and embryonic development (Karapetsas et al., 2011; Charron et al., 2012). U0126 (1,4-diamino-2,3-dicyano-1,4-bis[2-aminophenylthio]butadiene), a potent and specific inhibitor of MEK, has been used widely to determine MAPK function in various processes (Favata et al., 1998).
The S6K1 gene and protein have been examined extensively in human, mouse, and rat but not in goat, due to the lack of basic data for this animal. To study the expression and regulation of S6K1 in Inner Mongolia Cashmere goat cells, we cloned the full-length S6K1 cDNA, measured its expression in tissues by quantitative real-time PCR and immunohistochemistry, and investigated its activity in goat fetal fibroblasts with regard to its regulation by the MAPK signaling pathway.
MATERIALS AND METHODS
Animals and tissue collection
Inner Mongolia Cashmere goats were bred on natural semi-desert grassland and grazing in the pasture in Inner Mongolia, China. Brain, heart, liver, spleen, kidney, lung, and testis were collected from 5 adult male goats after slaughter in a commercial goat slaughter farm in the spring of 2012. Tissue samples were flash-frozen in liquid nitrogen immediately after harvest and stored at –80°C.
Cell cultures
Inner Mongolia Cashmere goat fetal fibroblasts (GFbs) were cultured in DMEM/F12 (Gibco) that was supplemented with 10% fetal bovine serum (FBS, HyClone Laboratories, Inc. Logan, UT USA), 100 U/ml penicillin G, and 100 mg/ml streptomycin (Sigma-Aldrich, Inc. St. Louis USA) and maintained as monolayer cultures at 37°C in humidified air with 5% CO2. Morphology was examined by light microscopy.
Reagents and antibodies
The MEK inhibitor U0126 was purchased from Sigma-Aldrich (Sigma-Aldrich, Inc. St. Louis USA) and dissolved in DMSO (Sigma-Aldrich). The concentration of DMSO in the final solution did not exceed 1% (v/v). The following primary antibodies were used: anti-β-actin (Sigma-Aldrich, Inc. St. Louis USA), anti-p70S6K (Santa Cruz Biotechnology, Inc. Santa Cruz, CA, USA), and anti-phosphorylated p70S6K (Thr389) (Cell Signaling Technology, Inc. Danvers, MA, USA).
RNA extraction and cDNA synthesis
Total RNA from brain, heart, liver, spleen, kidney, lung, and fetal fibroblasts of Inner Mongolia Cashmere goats was isolated using RNAzol (RNAiso Plus, TaKaRa Co. Ltd., China). RNA was reverse-transcribed with an oligo (dT)12–18 primer using the AMV 1st Strand cDNA Synthesis kit (Takara Co. Ltd., China) following the manufacturer’s procedure. Full-length S6K1 was cloned using cDNA1 from fetal fibroblasts, and cDNA2 from various tissues was quantified by real-time PCR. 3′RACE was performed with cDNA3, for which RNA was reverse-transcribed with reagents in the SMARTTM RACE cDNA Amplification Kit (Clontech, Laboratories, Inc. CA, USA). One microgram of total RNA was used for each reaction.
Cloning and sequencing of S6K1
The CDS fragment of the S6K1 gene was amplified by PCR for 35 cycles with cDNA1 as template at the appropriate annealing temperature for following primer pair: forward: 5′- ATGAGGCGACGAAGGAGGC -3′ and reverse: 5′- TCA CAGGTGCTCTGGTCGTTTG-3′; the predicted fragment length was 1,578 bp. The PCR products were electrophoresed, and photos were taken on a UV transilluminator (UVItec, UK). The amplicons were cloned into plasmids and sequenced on an ABI PRISM 377XL DNA Sequencer (Applied Biosystems, Inc. Foster City, CA, USA). The 3′ UTR fragment was amplified with cDNA3 as template for 30 cycles at 67°C using the following primer pairs: forward: 5′-GCCAGCACGGCAAATCCGCAGAC-3′; reverse (UPM): long (0.4 μM) 5′-CTAATACGACT CACTATAGGGCAAGCAGTGGTATCAACGCAGAGT-3′ and short (2 μM) 5′-CTAATACGAC TCACTATAGGGC-3′. 3′ RACE generated a single fragment of 932 bp.
Quantitative real-time PCR
The distribution of S6K1 mRNA between tissues was examined by quantitative real-time PCR. S6K1 cDNA was amplified with cDNA2 as template by real-time PCR using the following primers, forward: 5′ - AAATGCTGCTTCTC GTCTTG -3′ and reverse: 5′ - CTTGGAATCAAACTGA CTCACATC -3′; β-actin was amplified with forward: 5′ - CCACTGGCATTGTCATGGACTC -3′ and reverse: 5′ -TTCCTTGAT GTCACGGACGATTT -3′ primers.
Quantitative real-time PCR was performed on a Bio-Rad Chromo 4 PCR System using SYBR® Premix Ex TaqTM (Perfect Real Time) (TaKaRa Co. Ltd., China). One microliter of cDNA was amplified in a 25-® μl reaction, containing 10 μmol/L forward primer (0.5 μl), 10 μmol/L reverse primer (0.5 μl), 2×SYBR® Premix Ex TaqTM (12.5 μl), and nuclease-free water (10.5 μl). The following program was run: 95°C for 5 min; 40 cycles of 95°C for 15 s, 54°C for 30 s, and 72°C for 30 s; and 72°C for 10 min; a final melting curve was also generated. Three technical replicates were run. Delta CT (ΔCT) values were calculated to determine the expressions in the tissues. The real-time PCR results were analyzed by one-way analysis of variance (ANOVA) to compare expressions between tissues.
Immunohistochemistry
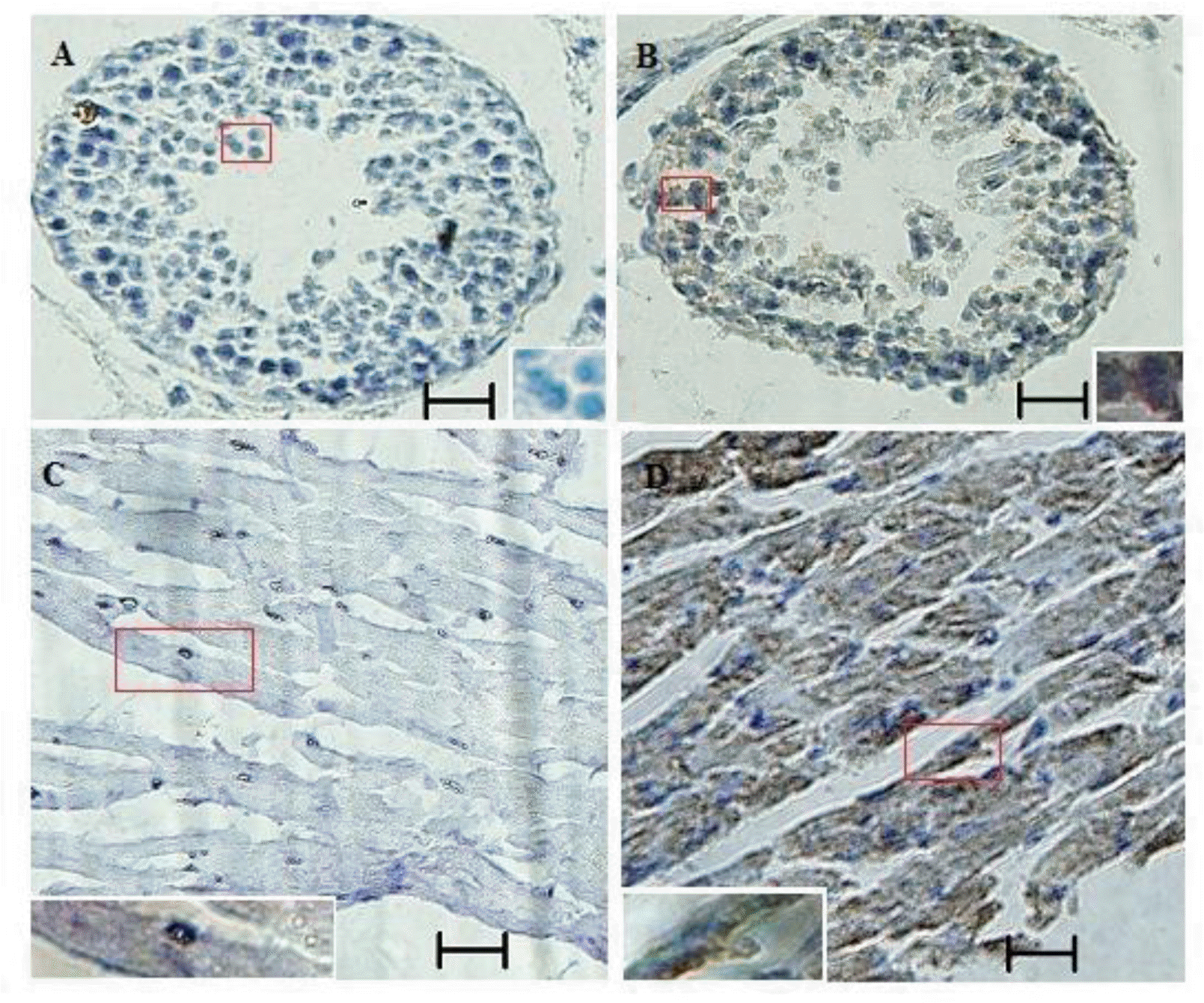
The tissue samples were fixed in 4% paraformaldehyde, dehydrated through an alcohol series, paraffin-embedded, and sectioned. Sections were deparaffinized in an alcohol gradient, blocked with normal nonimmune animal serum, incubated with rabbit anti-p70S6K (1:100) at 4°C overnight, and washed 3 times with PBS for 5 min. Then, the sections were incubated with biotinylated anti-rabbit IgG (1:50) for 10 min at room temperature, washed as before, and stained with DAB (3, 3′-diaminobenzidine tetrahydrochloride) using the UltraSebsitive TM SP kit (Maixin_Bio, Fuzhou, China) following the manufacturer’s instructions. Before being mounted, the sections were counterstained with hematoxylin. Sections of each tissue were immunostained without primary antibody and incubated in PBS as negative controls.
Western blot analysis
GFb cells were plated onto 9 cm2 dishes at 3×105 per dish and incubated. Subconfluent cells were treated with 1.5 μM of the MEK inhibitor U0126 for 1 h, collected with trypsin, washed 3 times with cold PBS, and lysed in buffer that contained 20 mM Tris (pH 8.0), 137 mM NaCl, 100 g/L glycerol, 50 g/L Triton X-100, 2 g/L Na2VO4, and 4 g/L EDTA; 10 ml PMSF (0.1 M) and 10 ml ALT (10 g/L) were added per 1 ml lysis buffer immediately before use. The cell lysates were put on ice for 15 min and centrifuged at 15,000 rpm at 4°C for 20 min, and the supernatant was transferred to new tubes.
The concentrations of the lysates were measured by Bio-Rad protein determination method (Bio-Rad Laboratories). Equal amounts (45 μg) of protein were electrophoresed on 12% (w/v) sodium dodecylsulfate polyacrylamide gels. Proteins were transferred to Hybond-polyvinylidene difluoride membranes (Amersham) and incubated with the primary antibodies overnight at 4°C and peroxidase-conjugated secondary antibodies at room temperature for 1 h. Enhanced chemiluminescence (ECL) (Amersham) was used to detect the signals.
Bioinformatics analysis
The nucleotide sequence of goat S6K1 cDNA and the deduced amino acid sequence were identified using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/). ORFs and the theoretical molecular weights of the deduced polypeptides were predicted using the Protein property calculator (http://www.basic.northwestern.edu/biotools/proteincalc.html). The protein’s isoelectric point was predicted using an online calculator (http://isoelectric.ovh.org/). Protein domains were analyzed using the SMART program (http://smart.embl-heidelberg.de/). Protein prosite patterns were identified using Psite (http://www.softberry.com). Models of certain regions were generated using SWISS-MODEL Workspace (Guex and Peitsch, 1997; Schwede et al., 2003; Arnold et al., 2006).
Statistical analysis
Descriptive statistics were generated for all quantitative data, expressed as mean±SD. Each assay was performed in triplicates.
RESULTS
Cloning and sequence analysis of Cashmere goat S6K1
S6K1 cDNA (GenBank accession GU144017.1) from the Inner Mongolia Cashmere goat comprises an open reading frame of 1,578 bp and 694 bp of 3′ untranslated region, with a polyadenylation signal at nucleotides 2,218–2,223 (AATAAA). The CDS sequence shares 99% identity with the bovine sequence and 94%, 92%, and 90% identity with human, rat, and mice sequence, respectively. The amino acid sequence is 99% identical to the bovine and human sequences.
Primary and secondary structure of Inner Mongolia Cashmere goat p70S6K
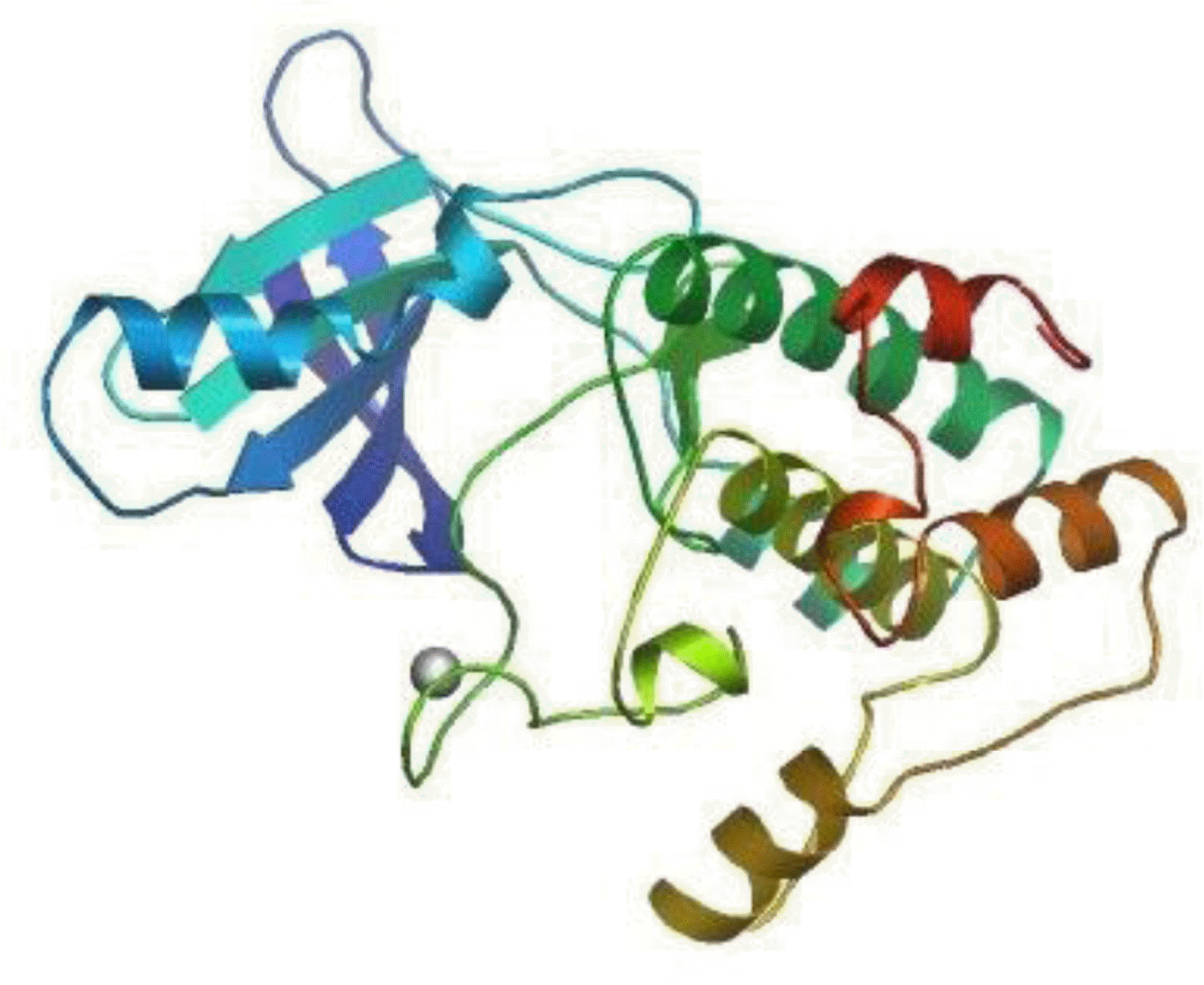
The deduced primary structure of the Inner Mongolia Cashmere goat p70S6K consists of 525 amino acids, and the predicted molecular weight is 59,251 Da for the unmodified protein; the estimated isoelectric point (pI) is 6.68. p70S6K comprises 14.09% acidic amino acids and 15.81% basic amino acids. p70S6K has an S_TKc domain (serine/threonine protein kinase, catalytic domain) from residues 91 to 352 and an extension to the Ser/Thr-type protein kinase domain from 353 to 415 (Figure 1A). There is 1 N-glycosylation site, 5 cAMP- and cGMP-dependent protein kinase phosphorylation sites, and 2 protein kinase C phosphorylation sites (Figure 1B). As calculated by SWISS-MODEL workspace, there is a catalytic domain from residues 91 to 352 in goat p70S6K; the quaternary structure is shown in Figure 2.
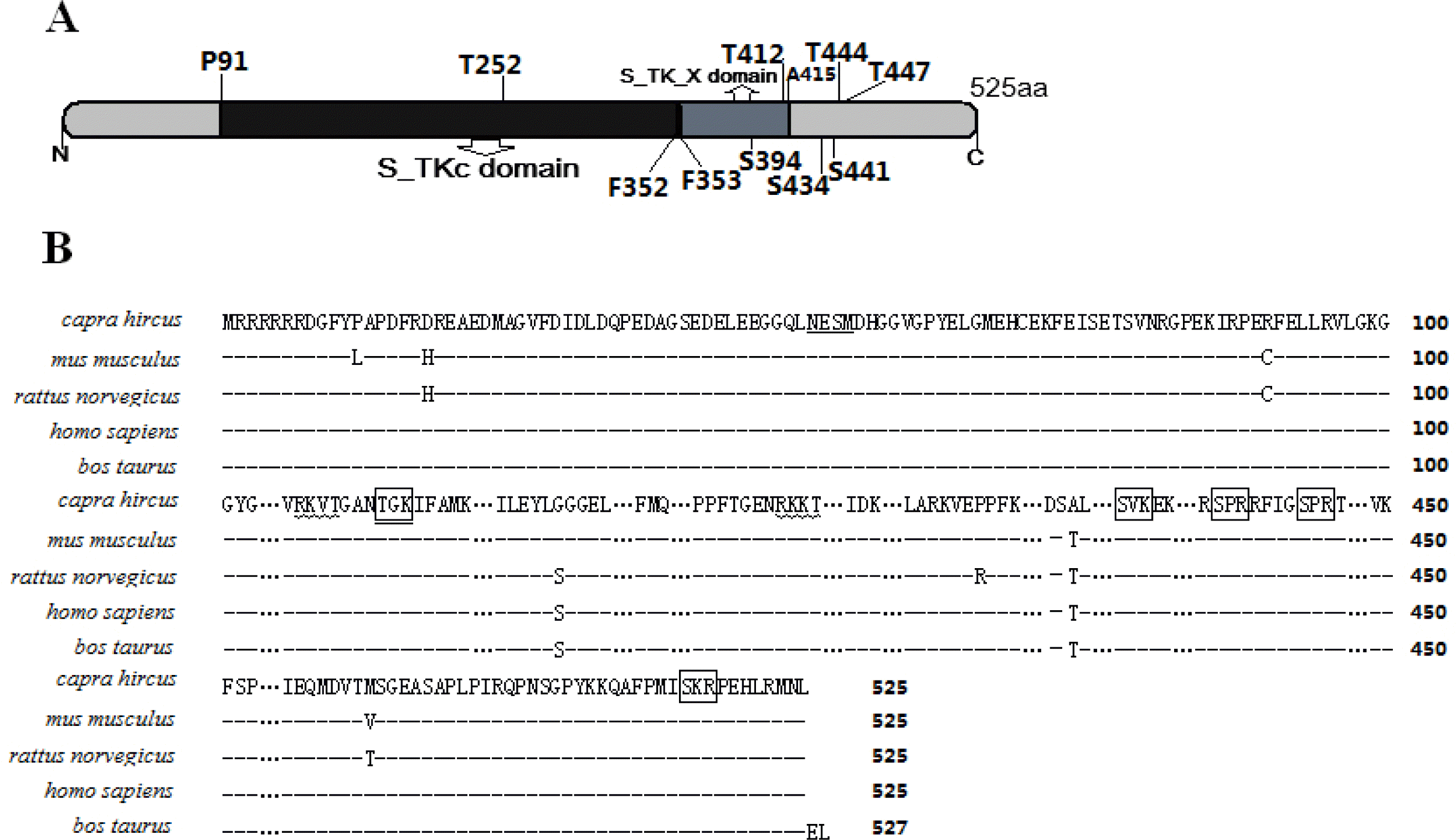
The predicted amino acid sequence and domains of p70S6K (A) There are two main domains in p70S6K which is 525 amino acids. The S_TKc domain is serine/threonine protein kinase catalytic domain. The S_TK_X domain is extended to Ser/Thr-type protein kinases, predicted with SMART (http://smart.embl.de/). (B) Deduced amino acid sequence of p70S6K. Alignment of the amino acid sequences of Capra hircus (GU-144017), Mus musculus (NP_001107806.1), Rattus norvegicus (NP_114191.1), Homo sapiens (NP_003152.1), and Bos taurus (NP_991385.1). Predicted N-glycolsylation sites are underlined. Protein kinase C phosphorylation sites are marked by billows, and cAMP- and cGMP-dependent protein kinase phosphorylation sites are marked by panes. All sites were determined using Psite (http://www.softberry.com).
Tissue distribution of Cashmere goat S6K1 mRNA and p70S6K protein
The relative abundance of S6K1 mRNA was assessed in brain, heart, liver, spleen, kidney, and lung tissue from Cashmere goat by quantitative real-time PCR. mRNA levels were higher in brain, heart, spleen, and lung compared with liver and kidney (Figure 3). p70S6K protein was detected in testis and heart of Inner Mongolia Cashmere goat by immunohistochemistry (Figure 4).
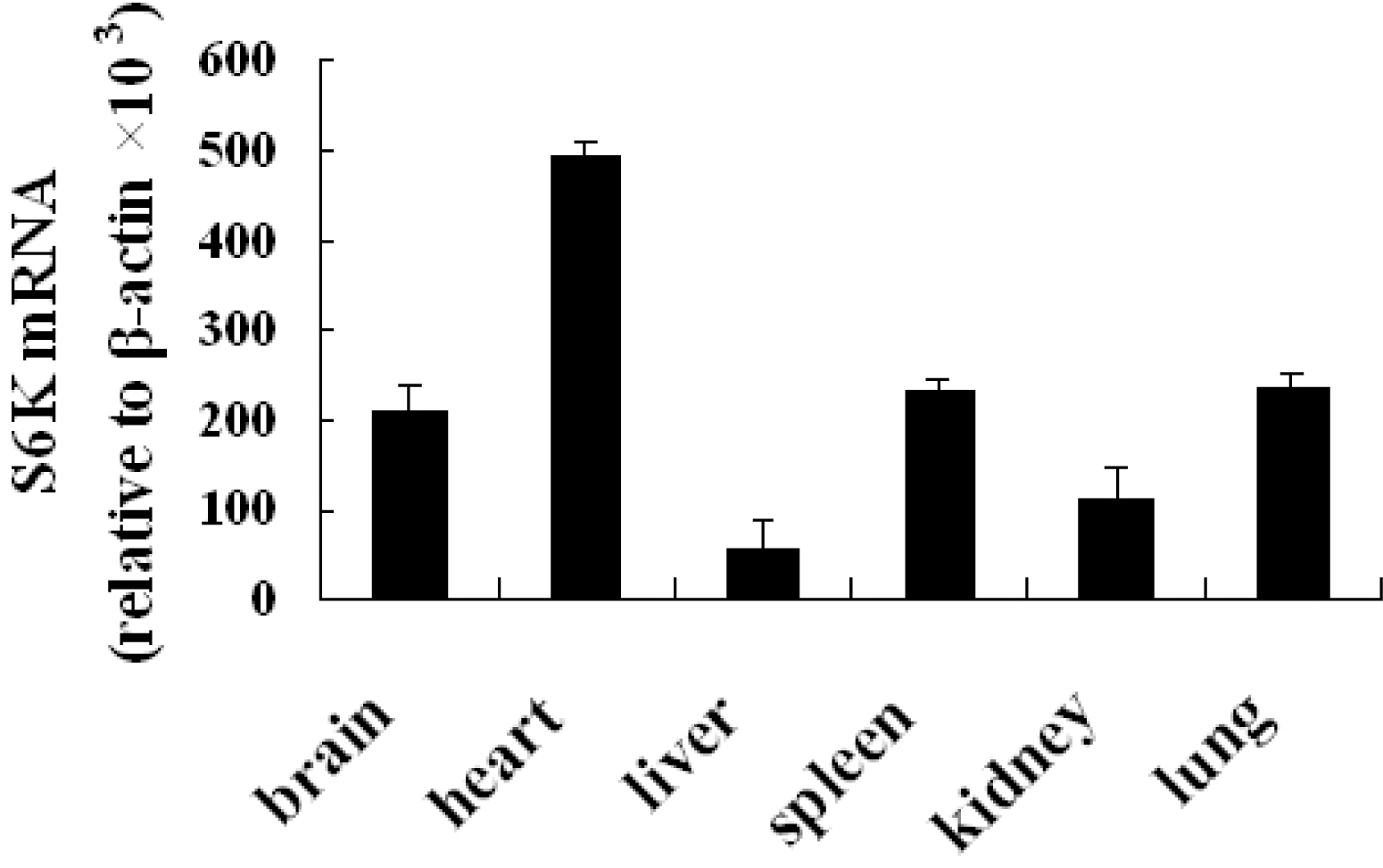
Quantitative real-time PCR analysis of S6K1 mRNA expressions in various tissues. mRNA levels are higher in heart compared with brain, liver, spleen, lung, and kidney.
U0126 inhibits phosphorylation of p70S6K
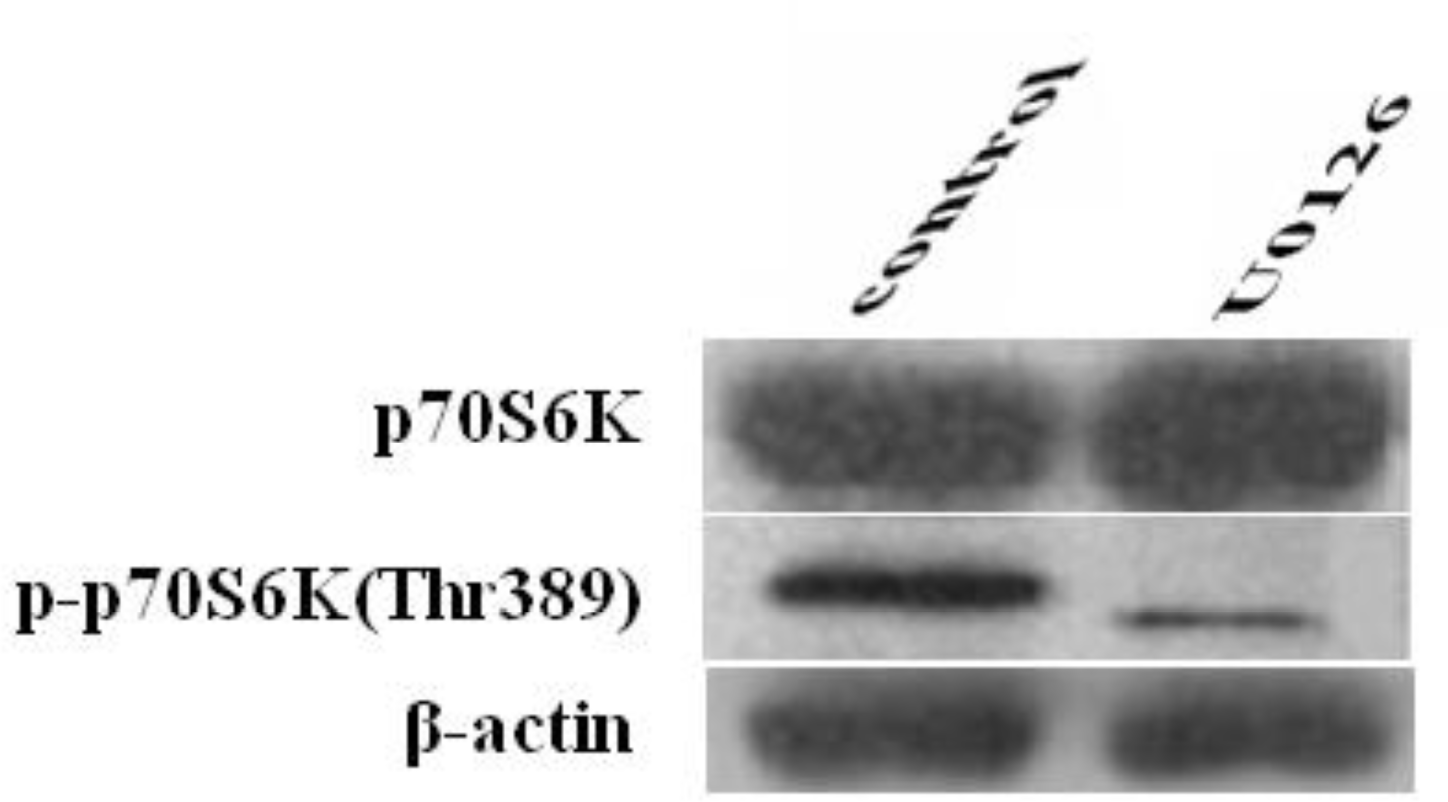
The expression of p70S6K and p-p70S6K (Thr389) was examined by western blot to study the inhibitory effects of U0126 on p70S6K and p-p70S6K (Thr389) expression. As shown in Figure 5, the expression of p-p70S6K (Thr389) in Cashmere goat fetal fibroblasts was inhibited by treatment with U0126 for 1 h, whereas that of p70S6K was unaffected, suggesting that phosphorylation of p70S6K (Thr389) was prevented by an inhibitor that targets MEK. We can conclude that phosphorylation of p70S6K (Thr389) is regulated by MAPK signaling.
DISCUSSION
S6K1 lies on chromosome 17 in humans, and its CDS encodes two isoforms. Translation of the longer species (isoform 1, p85S6K) is initiated at Met1, generating a protein that is 525 amino acid residues, whereas translation of the shorter isoform (isoform 2, p70S6K) is initiated at Met24, yielding 502 amino acid residues (GenBank accession number NC_000017). S6K1 localizes to chromosome 19 in bovine, and its product, p70S6K, consists of 527 amino acids (GenBank accession number NP_991385).
In this study, we examined S6K1 in the Inner Mongolia Cashmere goat. The ORF of goat S6K1 cDNA encodes 525 amino acids, and by bioinformatics analysis, there was no nuclear localization sequence (NLS). In the deduced amino acid sequence of goat p70S6K, there is a catalytic kinase domain between residues 91 to 352, similar to other reported AGC kinase family members (Proud, 2002; Brian et al., 2012).
Of the two known isoforms, p85S6K is believed to target to the nucleus, whereas p70S6K is primarily cytoplasmic (Kim et al., 2009). Nevertheless, despite the absence of a classical NLS, p70S6K was nucleocytoplasmic in location and can shuttle from the cytoplasm to the nucleus on growth factor stimulation (Valovka et al., 2003; Rosner and Hengstschläger, 2011).
Nuclear localization of p70S6K depends on mTOR-mediated phosphorylation at T389 (Rosner and Hengstschläger, 2011; Rosner et al., 2012). mTORC1 regulates p70S6K activity and its localization, suggesting that p70S6K is in the cytoplasm and the nucleus, regulating transcription. Based on existing data, mTOR-dependent regulation of transcription requires p70S6K (Hannan et al., 2003; Düvel et al., 2010; Li et al., 2011), but the mechanisms by which p70S6K executes its functions in the nucleus are unknown.
p70S6K is a serine/threonine kinase that is activated in response to growth factors, cytokines, and nutrients. It has been linked to many cellular processes, including protein synthesis, mRNA processing, glucose homeostasis, cell growth, and survival (Fenton and Gout, 2011).
In this study, we detected S6K1 in certain goat tissues by real-time PCR (Figure 3) and immunohistochemistry (Figure 4) in a pattern that is similar to that in humans (Gout et al., 1998). The lack of basic data on genes and their expressions in goat has impeded molecular studies in this important livestock. Our study provides novel findings on S6K1 of cashmere goats, which might be valuable to researchers of goat and sheep.
p70S6K is the best-defined substrate of mTOR (Chung et al., 1992; Hay and Sonenberg, 2004). S6K is suppressed by the mTOR inhibitor RAD001 in SEG-1 esophageal cancer cells (Wang et al., 2010) and by CCI-779 in goat fetal fibroblasts (Wang et al., 2012). MEK are upstream regulators of mTOR, and can mediate amino acid signal-induced phosphorylation of S6K1 in HEK293 cells (Casas-Terradellas et al., 2008). U0126, an inhibitor of the MEK/ERK pathway, abolishes insulin-induced S6K1 phosphorylation and activity in chicken myoblasts (Duchêne et al., 2008).
Nevertheless, when human urothelial carcinoma cells are treated with U0126, ERK1/2 phosphorylation is absent, but p70S6K phosphorylation is unchanged (Nawroth et al., 2011). This phenomenon has never been investigated in domestic species. Thus, we examined the signal transduction pathway that mediates S6K1 activation by inhibiting the function of MEK in goat fetal fibroblasts. Our data demonstrate that the phosphorylation of p70S6K (Thr389) is inhibited by U0126 (Figure 5), indicating that significant controversy remains regarding the function of MEK in the activation of p70S6K.
Ribosomal protein S6 (rpS6) is a well-characterized downstream molecule that is regulated by p70S6K, and for at least 10 yrs after the discovery of S6K, rpS6 was believed to be its only substrate. In recent years, several S6K1 substrates that control protein synthesis, cytoskeletal rearrangement, proliferation, splicing, feedback regulation, and cell survival have been identified (Fenton and Gout, 2011). Many functional aspects of S6K1 have been characterized-mTOR-mediated phosphorylation of p70S6K at T389 also regulates its nucleocytoplasmic localization (Rosner et al., 2012), crosstalk exists between the mTOR/S6K1 and Hedgehog (HH) pathways (Wang et al., 2012), and mTORC1-activated p70S6K phosphorylates Rictor on T1135 and regulates mTORC2 signaling (Dibble et al., 2009; Julien et al., 2010). These findings provide significant insights into the regulation of p70S6K and have increased our understanding of its function.
In this study, we examined S6K1 and its expression in Cashmere goats, demonstrating that it is expressed in the indicated goat tissues and cells. We infer that activation of p70S6K is regulated by MAPK signaling, but further studies are needed to determine how p70S6K functions in Inner Mongolia Cashmere goat cells.
CONCLUSIONS
S6K1 cDNA cloned from Inner Mongolia Cashmere goats, comprised of 2,272 bp and included an ORF of 1,578 bp-corresponding to a polypeptide of 525 amino acids and a 3′UTR of 694 bp. S6K1 is expressed at higher levels in brain, heart, spleen, and lung compared with kidney and liver. The phosphorylation of p70S6K is regulated by MAPK signaling.
Acknowledgements
This work was supported by a grant from the Natural Sciences Foundation of China (No. 31160469) and Natural Sciences Foundation of Inner Mongolia (2011MS0521).