 |
 |
Abstract
ACKNOWLEDGMENTS
Figure 1.
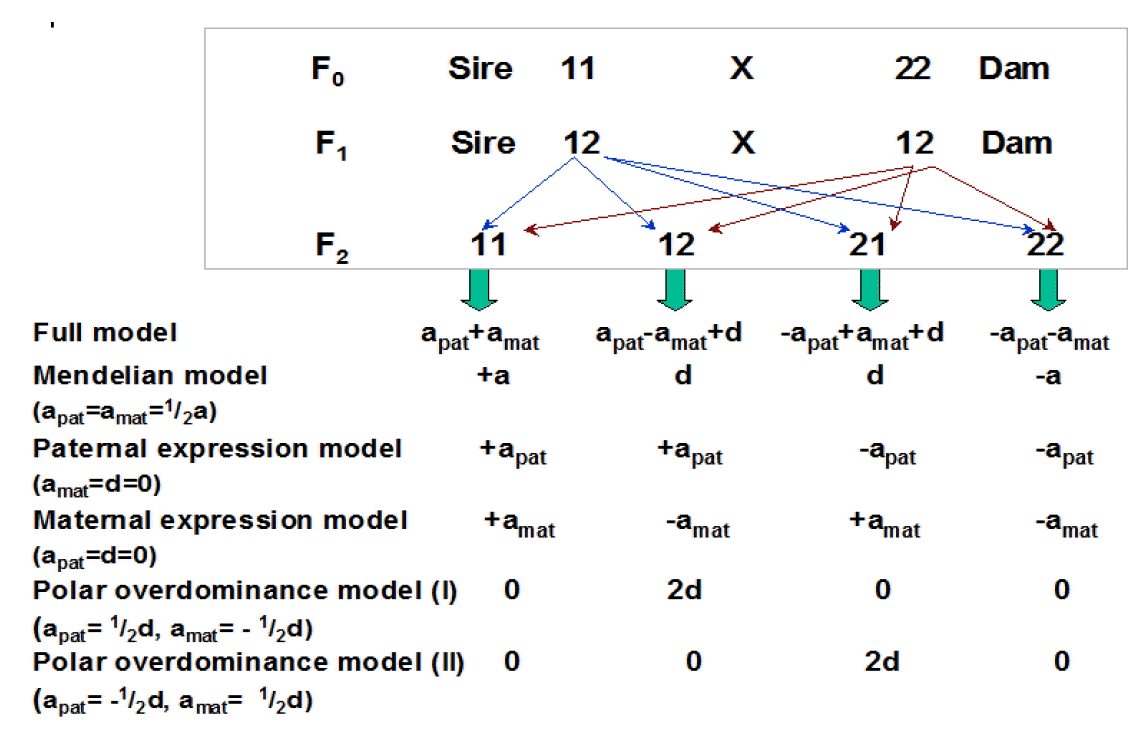
Table 1.
| QTL inheritance mode | Genetic variance |
QTL effecta
|
||
|---|---|---|---|---|
| Large | Medium | Small | ||
| Additive (a) | 0.5a2 | 0.800 | 0.500 | 0.320 |
| Complete dominance (a = d) | 0.5a2+0.25d2 | 0.654 | 0.408 | 0.261 |
| Parental-origin expression (a) | a2 | 0.566 | 0.354 | 0.226 |
| Polar overdominance (d) | 0.75d2 | 0.654 | 0.408 | 0.261 |
a Different QTL effects under varying genetic models were set such that large, medium and small QTL explained 32%, 12.5% or 5.1%, respectively, of the phenotypic variance. Error variances were set 0.680, 0.875 and 0.949 for large, medium and small QTL, respectively, such that overall phenotypic variances become standard unit (1.0).
Table 2.
| POD QTL effect (allele frequency)a | Power to detect QTL (%)b | Declared POD type (%)c | Declared non-POD type (%)d | POD QTL position (true = 75 cM)e | POD QTL effect (d)f | POD QTL variance (%)g | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||||||||
| Mend | Pat Exp | Mat Exp | Pod I | Pod II | I | II | both | Mend | Pat Exp | Mat Exp | Partial Exp | ||||
| Design I (N = 512) | |||||||||||||||
| I 0.65 (1.0) | 100 | 100 | 100 | 100 | 100 | 99 | 0 | 0 | 0 | 0 | 0 | 100 | 75.0 (1.6) | 0.65 (0.05) | 32.2 (5.0) |
| I 0.65 (0.8) | 63 | 79 | 77 | 90 | 82 | 59 | 3 | 6 | 2 | 10 | 8 | 69 | 74.2 (9.2) | 0.39 (0.14) | 13.1 (8.4) |
| I 0.41 (1.0) | 90 | 97 | 98 | 100 | 76 | 93 | 0 | 6 | 0 | 4 | 4 | 92 | 75.0 (3.4) | 0.41 (0.05) | 12.6 (3.2) |
| I 0.41 (0.8) | 36 | 55 | 57 | 76 | 48 | 54 | 4 | 10 | 3 | 10 | 12 | 50 | 73.3 (11.9) | 0.27 (0.09) | 6.1 (3.3) |
| I 0.26 (1.0) | 42 | 63 | 66 | 99 | 37 | 83 | 1 | 12 | 7 | 17 | 17 | 55 | 74.8 (7.1) | 0.27 (0.05) | 5.5 (2.2) |
| I 0.26 (0.8) | 16 | 30 | 33 | 56 | 26 | 41 | 4 | 9 | 7 | 6 | 9 | 28 | 72.6 (15.6) | 0.21 (0.07) | 3.6 (1.6) |
| II 0.65 (1.0) | 100 | 100 | 100 | 100 | 100 | 0 | 98 | 1 | 0 | 0 | 0 | 100 | 74.9 (1.6) | 0.65 (0.05) | 31.9 (4.8) |
| II 0.65 (0.8) | 62 | 76 | 79 | 79 | 90 | 2 | 59 | 8 | 3 | 9 | 11 | 67 | 74.5 (8.5) | 0.38 (0.15) | 12.4 (8.8) |
| II 0.41 (1.0) | 91 | 97 | 97 | 79 | 100 | 0 | 89 | 10 | 0 | 2 | 3 | 95 | 74.9 (3.3) | 0.41 (0.05) | 12.9 (3.2) |
| II 0.41 (0.8) | 34 | 58 | 55 | 50 | 79 | 5 | 57 | 11 | 3 | 12 | 10 | 50 | 74.7 (9.8) | 0.27 (0.09) | 6.2 (3.7) |
| II 0.26 (1.0) | 43 | 68 | 66 | 42 | 99 | 1 | 81 | 16 | 3 | 17 | 18 | 59 | 74.3 (7.2) | 0.27 (0.05) | 5.7 (2.0) |
| II 0.26 (0.8) | 16 | 36 | 26 | 26 | 57 | 6 | 42 | 7 | 5 | 8 | 5 | 30 | 73.3 (14.9) | 0.21 (0.07) | 3.7 (1.8) |
| Design I (N = 1024) | |||||||||||||||
| I 0.41 (1.0) | 99 | 100 | 100 | 100 | 97 | 93 | 0 | 2 | 0 | 0 | 0 | 100 | 75.0 (1.9) | 0.41 (0.04) | 12.6 (2.4) |
| I 0.41 (0.8) | 53 | 76 | 72 | 88 | 75 | 51 | 4 | 6 | 2 | 11 | 7 | 69 | 74.2 (9.2) | 0.25 (0.09) | 5.2 (3.2) |
| I 0.26 (1.0) | 81 | 94 | 92 | 100 | 68 | 88 | 0 | 8 | 0 | 4 | 6 | 89 | 74.9 (4.1) | 0.26 (0.04) | 5.3 (1.5) |
| I 0.26 (0.8) | 24 | 45 | 51 | 69 | 44 | 46 | 7 | 6 | 4 | 10 | 13 | 40 | 74.4 (10.3) | 0.18 (0.05) | 2.6 (1.4) |
| II 0.41 (1.0) | 100 | 100 | 100 | 96 | 100 | 0 | 91 | 2 | 0 | 1 | 0 | 99 | 74.9 (2.0) | 0.41 (0.04) | 12.6 (2.3) |
| II 0.41 (0.8) | 50 | 71 | 74 | 68 | 86 | 3 | 52 | 4 | 2 | 9 | 12 | 63 | 74.1 (8.1) | 0.25 (0.09) | 5.2 (3.4) |
| II 0.26 (1.0) | 81 | 95 | 90 | 68 | 100 | 0 | 90 | 7 | 0 | 6 | 7 | 87 | 75.1 (4.3) | 0.26 (0.04) | 5.3 (1.4) |
| II 0.26 (0.8) | 26 | 53 | 53 | 44 | 73 | 4 | 52 | 7 | 5 | 12 | 12 | 45 | 72.5 (12.0) | 0.18 (0.05) | 2.7 (1.5) |
| Design II (N = 513) | |||||||||||||||
| I 0.65 (1.0) | 100 | 100 | 100 | 100 | 100 | 95 | 0 | 1 | 0 | 0 | 0 | 100 | 74.9 (1.5) | 0.65 (0.05) | 32.0 (4.5) |
| I 0.65 (0.8) | 47 | 88 | 88 | 99 | 88 | 65 | 3 | 6 | 0 | 9 | 10 | 79 | 74.7 (6.1) | 0.35 (0.09) | 9.8 (4.9) |
| I 0.41 (1.0) | 87 | 98 | 97 | 100 | 79 | 92 | 1 | 5 | 0 | 3 | 4 | 93 | 75.1 (3.5) | 0.41 (0.06) | 12.6 (3.4) |
| I 0.41 (0.8) | 20 | 56 | 61 | 90 | 48 | 63 | 4 | 10 | 2 | 12 | 17 | 52 | 74.0 (10.3) | 0.24 (0.06) | 4.5 (2.2) |
| I 0.26 (1.0) | 47 | 64 | 66 | 99 | 36 | 84 | 0 | 9 | 6 | 15 | 17 | 58 | 74.3 (6.6) | 0.27 (0.05) | 5.5 (2.0) |
| I 0.26 (0.8) | 10 | 22 | 30 | 51 | 24 | 35 | 8 | 5 | 6 | 6 | 5 | 22 | 72.8 (16.4) | 0.19 (0.04) | 2.9 (1.1) |
| II 0.65 (1.0) | 100 | 100 | 100 | 100 | 100 | 0 | 94 | 1 | 0 | 0 | 0 | 100 | 75.0 (1.4) | 0.65 (0.05) | 32.2 (4.6) |
| II 0.65 (0.8) | 45 | 90 | 93 | 88 | 99 | 2 | 67 | 5 | 0 | 6 | 10 | 83 | 74.7 (5.7) | 0.35 (0.09) | 10.0 (4.9) |
| II 0.41 (1.0) | 90 | 97 | 97 | 75 | 100 | 0 | 92 | 4 | 0 | 2 | 2 | 96 | 75.1 (3.6) | 0.41 (0.05) | 12.7 (3.2) |
| II 0.41 (0.8) | 20 | 57 | 61 | 48 | 86 | 4 | 63 | 10 | 3 | 12 | 14 | 49 | 74.2 (10.1) | 0.24 (0.06) | 4.5 (2.2) |
| II 0.26 (1.0) | 40 | 65 | 69 | 43 | 99 | 0 | 82 | 11 | 4 | 17 | 19 | 57 | 75.0 (6.7) | 0.27 (0.05) | 5.6 (2.0) |
| II 0.26 (0.8) | 9 | 30 | 27 | 23 | 55 | 8 | 37 | 6 | 5 | 6 | 5 | 29 | 72.0 (16.8) | 0.19 (0.05) | 2.9 (1.1) |
| Design II (N = 1026) | |||||||||||||||
| I 0.41 (1.0) | 100 | 100 | 100 | 100 | 97 | 94 | 0 | 2 | 0 | 0 | 0 | 100 | 74.9 (1.8) | 0.41 (0.04) | 12.6 (2.2) |
| I 0.41 (0.8) | 41 | 84 | 82 | 96 | 81 | 65 | 1 | 9 | 1 | 13 | 12 | 71 | 74.8 (5.6) | 0.22 (0.05) | 4.0 (1.9) |
| I 0.26 (1.0) | 79 | 92 | 91 | 100 | 69 | 86 | 0 | 8 | 0 | 7 | 8 | 85 | 75.3 (3.8) | 0.26 (0.04) | 5.2 (1.6) |
| I 0.26 (0.8) | 19 | 53 | 53 | 84 | 39 | 60 | 5 | 7 | 4 | 14 | 15 | 42 | 74.1 (9.8) | 0.16 (0.04) | 2.1 (0.9) |
| II 0.41 (1.0) | 100 | 100 | 100 | 97 | 100 | 0 | 96 | 1 | 0 | 0 | 0 | 100 | 75.1 (1.6) | 0.41 (0.04) | 12.6 (2.3) |
| II 0.41 (0.8) | 36 | 80 | 85 | 78 | 97 | 3 | 68 | 10 | 0 | 12 | 12 | 72 | 74.7 (6.9) | 0.22 (0.05) | 3.8 (1.8) |
| II 0.26 (1.0) | 82 | 94 | 93 | 67 | 100 | 0 | 88 | 6 | 0 | 6 | 6 | 89 | 74.9 (4.2) | 0.26 (0.04) | 5.3 (1.5) |
| II 0.26 (0.8) | 17 | 49 | 51 | 43 | 84 | 5 | 58 | 10 | 4 | 11 | 11 | 45 | 73.8 (13.1) | 0.16 (0.04) | 2.0 (0.9) |
Polar overdominance (POD) QTL were simulated based on different allele frequencies, magnitudes, and number of F2 progeny in two mating designs I: 2 F0 sires, 10 F0 dams, 8 F1 sires for 512 or 1,024 F2 progeny; II: 20 F0 sires, 80 F0 dams, 19 F1 sires for 513 or 1,026 F2 progeny. A biallelic QTL was simulated at 75 cM for a 100 cM chromosome with 11 equidistant markers. Each marker had four alleles with different allele frequencies in parental breeds (0.6 (0.1), 0.2 (0.1), 0.1 (0.2), 0.1 (0.6) in breed A (B)). A total of 500 replicates were generated per each parameter set.
a POD I or II refer to QTL, for which differential phenotype is observed for QTL genotype 12 (I) against genotype effects of 11, 21, and 22, or genotype 21 (II) against 11, 12, and 22. POD QTL effects (d), 0.65, 0.41, and 0.26 were defined as large, medium or small QTL, such that the QTL explained 32%, 12.5% or 5.1%, respectively, of the phenotypic variance. Alternate QTL alleles were homogenously (1.0/0.0) or differently (0.8/0.2) distributed in F0 parental breeds.
b Proportion of replicates in which a POD QTL was detected at a 5% chromosome-wise level in Mendelian, paternal expression, maternal expression, POD I, or POD II models.
c If a POD QTL was detected at a 5% chromosome-wise level in its respective POD model (Test1), a series of POD tests (Tests 2 and 3) were performed for the POD QTL to be declared as POD I, POD II, or both types.
d POD QTL were tested with a series of tests to differentiate parent-of-origin effects (paternal, maternal, or partial expression) from Mendelian effects, according to THOMSEN et al. (2004).
e Mean estimates (standard deviations) of the most likely QTL position from the replicates with POD QTL evidence in respective POD models.
Table 3.
| QTL effect (allele frequency)a | Power to detect QTL (%)b | Declared POD type (%)c | QTL effect (allele frequency)a | Power to detect QTL (%)b | Declared POD type (%)c | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||||||||||||||
| Mend | Pat Expr | Mat Expr | Pod I | Pod II | I | II | Both | None | Mend | Pat Exp | Mat Exp | Pod I | Pod II | I | II | Both | None | ||
| Design I (N = 512) | |||||||||||||||||||
| Pat 0.57 (1.0) | 100 | 100 | 4 | 100 | 100 | 0 | 0 | 0 | 100 | Add 0.80 (1.0) | 100 | 100 | 100 | 5 | 3 | 0 | 0 | 0 | 100 |
| Pat 0.57 (0.8) | 84 | 90 | 6 | 83 | 82 | 3 | 3 | 1 | 94 | Add 0.80 (0.8) | 91 | 85 | 89 | 16 | 13 | 3 | 2 | 0 | 95 |
| Pat 0.35 (1.0) | 100 | 100 | 5 | 98 | 98 | 1 | 2 | 0 | 97 | Add 0.50 (1.0) | 100 | 100 | 100 | 6 | 4 | 1 | 0 | 0 | 99 |
| Pat 0.35 (0.8) | 65 | 84 | 5 | 64 | 60 | 8 | 6 | 2 | 84 | Add 0.50 (0.8) | 82 | 71 | 74 | 10 | 9 | 2 | 2 | 0 | 96 |
| Pat 0.23 (1.0) | 78 | 99 | 4 | 65 | 64 | 10 | 9 | 2 | 79 | Add 0.32 (1.0) | 98 | 87 | 88 | 6 | 6 | 1 | 2 | 0 | 97 |
| Pat 0.23 (0.8) | 32 | 68 | 4 | 39 | 36 | 11 | 10 | 2 | 77 | Add 0.32 (0.8) | 57 | 47 | 43 | 8 | 7 | 3 | 3 | 0 | 94 |
| Mat 0.57 (1.0) | 100 | 3 | 100 | 100 | 100 | 0 | 0 | 0 | 100 | Dom 0.65 (1.0) | 100 | 100 | 100 | 89 | 85 | 0 | 0 | 0 | 100 |
| Mat 0.57 (0.8) | 86 | 5 | 93 | 84 | 84 | 4 | 2 | 0 | 93 | Dom 0.65 (0.8) | 92 | 82 | 82 | 39 | 37 | 2 | 1 | 0 | 97 |
| Mat 0.35 (1.0) | 100 | 4 | 100 | 98 | 98 | 1 | 1 | 0 | 98 | Dom 0.41 (1.0) | 100 | 98 | 97 | 42 | 45 | 1 | 1 | 0 | 98 |
| Mat 0.35 (0.8) | 64 | 4 | 82 | 58 | 57 | 7 | 6 | 1 | 85 | Dom 0.41 (0.8) | 76 | 59 | 54 | 17 | 12 | 3 | 2 | 0 | 94 |
| Mat 0.23 (1.0) | 72 | 5 | 99 | 68 | 64 | 11 | 10 | 3 | 76 | Dom 0.26 (1.0) | 99 | 65 | 67 | 18 | 19 | 2 | 1 | 0 | 97 |
| Mat 0.23 (0.8) | 34 | 4 | 64 | 30 | 31 | 9 | 7 | 2 | 82 | Dom 0.26 (0.8) | 48 | 33 | 27 | 9 | 7 | 3 | 2 | 0 | 94 |
| Design I (N = 1,024) | |||||||||||||||||||
| Pat 0.35 (1.0) | 100 | 100 | 5 | 100 | 100 | 0 | 0 | 0 | 100 | Add 0.50 (1.0) | 100 | 100 | 100 | 4 | 5 | 0 | 0 | 0 | 100 |
| Pat 0.35 (0.8) | 81 | 91 | 4 | 76 | 77 | 2 | 3 | 0 | 95 | Add 0.50 (0.8) | 92 | 84 | 85 | 9 | 10 | 1 | 1 | 0 | 98 |
| Pat 0.23 (1.0) | 97 | 100 | 5 | 91 | 94 | 1 | 1 | 0 | 98 | Add 0.32 (1.0) | 100 | 99 | 99 | 6 | 6 | 1 | 1 | 0 | 99 |
| Pat 0.23 (0.8) | 62 | 80 | 5 | 53 | 49 | 7 | 5 | 2 | 86 | Add 0.32 (0.8) | 78 | 66 | 66 | 6 | 11 | 1 | 3 | 0 | 95 |
| Mat 0.35 (1.0) | 100 | 4 | 100 | 100 | 100 | 0 | 0 | 0 | 100 | Dom 0.41 (1.0) | 100 | 100 | 100 | 76 | 72 | 0 | 0 | 0 | 100 |
| Mat 0.35 (0.8) | 83 | 7 | 93 | 77 | 79 | 3 | 3 | 1 | 93 | Dom 0.41 (0.8) | 87 | 76 | 75 | 24 | 25 | 2 | 2 | 0 | 96 |
| Mat 0.23 (1.0) | 99 | 6 | 100 | 94 | 95 | 1 | 1 | 0 | 98 | Dom 0.26 (1.0) | 100 | 94 | 90 | 30 | 37 | 1 | 0 | 0 | 98 |
| Mat 0.23 (0.8) | 58 | 7 | 82 | 52 | 57 | 5 | 5 | 2 | 88 | Dom 0.26 (0.8) | 69 | 47 | 46 | 13 | 14 | 3 | 2 | 0 | 95 |
| Design II (N = 513) | |||||||||||||||||||
| Pat 0.57 (1.0) | 100 | 100 | 5 | 100 | 100 | 0 | 0 | 0 | 100 | Add 0.80 (1.0) | 100 | 100 | 100 | 4 | 3 | 0 | 0 | 0 | 100 |
| Pat 0.57 (0.8) | 92 | 99 | 6 | 92 | 92 | 3 | 3 | 0 | 95 | Add 0.80 (0.8) | 100 | 96 | 99 | 8 | 9 | 0 | 0 | 0 | 99 |
| Pat 0.35 (1.0) | 99 | 100 | 4 | 98 | 98 | 0 | 0 | 0 | 99 | Add 0.50 (1.0) | 100 | 100 | 100 | 5 | 5 | 0 | 1 | 0 | 99 |
| Pat 0.35 (0.8) | 64 | 92 | 5 | 61 | 65 | 6 | 8 | 2 | 84 | Add 0.50 (0.8) | 91 | 74 | 76 | 6 | 6 | 1 | 1 | 0 | 97 |
| Pat 0.23 (1.0) | 90 | 100 | 4 | 80 | 82 | 4 | 4 | 0 | 92 | Add 0.32 (1.0) | 97 | 82 | 86 | 4 | 4 | 1 | 1 | 0 | 97 |
| Pat 0.23 (0.8) | 42 | 80 | 7 | 41 | 39 | 9 | 8 | 2 | 82 | Add 0.32 (0.8) | 59 | 43 | 41 | 9 | 5 | 4 | 2 | 0 | 94 |
| Mat 0.57 (1.0) | 100 | 5 | 100 | 100 | 100 | 0 | 0 | 0 | 100 | Dom 0.65 (1.0) | 100 | 100 | 100 | 87 | 88 | 0 | 0 | 0 | 100 |
| Mat 0.57 (0.8) | 95 | 4 | 99 | 92 | 93 | 1 | 2 | 0 | 97 | Dom 0.65 (0.8) | 98 | 89 | 89 | 25 | 21 | 1 | 1 | 0 | 98 |
| Mat 0.35 (1.0) | 100 | 4 | 100 | 98 | 98 | 1 | 1 | 0 | 98 | Dom 0.41 (1.0) | 100 | 98 | 99 | 43 | 38 | 0 | 1 | 0 | 99 |
| Mat 0.35 (0.8) | 64 | 5 | 95 | 64 | 64 | 7 | 9 | 2 | 82 | Dom 0.41 (0.8) | 84 | 56 | 64 | 11 | 12 | 1 | 3 | 0 | 96 |
| Mat 0.23 (1.0) | 88 | 6 | 100 | 82 | 86 | 4 | 5 | 1 | 91 | Dom 0.26 (1.0) | 98 | 69 | 65 | 19 | 19 | 1 | 3 | 0 | 96 |
| Mat 0.23 (0.8) | 39 | 4 | 81 | 38 | 35 | 9 | 10 | 3 | 78 | Dom 0.26 (0.8) | 48 | 26 | 31 | 7 | 6 | 3 | 2 | 0 | 95 |
| Design II (N = 1,026) | |||||||||||||||||||
| Pat 0.35 (1.0) | 100 | 100 | 5 | 100 | 100 | 0 | 0 | 0 | 100 | Add 0.50 (1.0) | 100 | 100 | 100 | 4 | 4 | 0 | 0 | 0 | 100 |
| Pat 0.35 (0.8) | 89 | 99 | 4 | 86 | 86 | 2 | 4 | 0 | 93 | Add 0.50 (0.8) | 100 | 100 | 100 | 6 | 4 | 0 | 0 | 0 | 100 |
| Pat 0.23 (1.0) | 98 | 100 | 4 | 96 | 95 | 1 | 1 | 0 | 98 | Add 0.32 (1.0) | 100 | 100 | 99 | 5 | 4 | 0 | 0 | 0 | 99 |
| Pat 0.23 (0.8) | 57 | 89 | 7 | 53 | 52 | 7 | 8 | 2 | 83 | Add 0.32 (0.8) | 86 | 68 | 67 | 4 | 6 | 1 | 1 | 0 | 98 |
| Mat 0.35 (1.0) | 100 | 5 | 100 | 100 | 100 | 0 | 0 | 0 | 100 | Dom 0.41 (1.0) | 100 | 100 | 100 | 75 | 74 | 0 | 0 | 0 | 100 |
| Mat 0.35 (0.8) | 93 | 5 | 100 | 89 | 92 | 3 | 2 | 0 | 95 | Dom 0.41 (0.8) | 98 | 85 | 85 | 18 | 17 | 1 | 1 | 0 | 99 |
| Mat 0.23 (1.0) | 98 | 4 | 100 | 94 | 95 | 1 | 1 | 0 | 97 | Dom 0.26 (1.0) | 100 | 93 | 93 | 35 | 31 | 0 | 1 | 0 | 99 |
| Mat 0.23 (0.8) | 59 | 4 | 91 | 53 | 49 | 11 | 6 | 1 | 82 | Dom 0.26 (0.8) | 76 | 49 | 49 | 10 | 10 | 2 | 2 | 0 | 96 |
Mendelian or parent-of-origin QTL were simulated based on different allele frequencies, magnitudes, and number of F2 progeny in two mating designs I and II (see Table 2).
a Add: QTL with additive effect only (d = 0), Dom: QTL with complete dominance (a = d), Pat: QTL with paternal expression, Mat: QTL with maternal expression. QTL effects for each type of inheritance were defined, in magnitude order, as large, medium or small QTL, such that the QTL explained 32%, 12.5% or 5.1%, respectively, of the phenotypic variance. Alternate QTL alleles were homogenously (1.0/0.0) or differently (0.8/0.2) distributed in F0 parental breeds.
REFERENCES
- TOOLS
-
METRICS

-
- 0 Crossref
- 0 Scopus
- 6,859 View
- 75 Download
- Related articles
-
Potential of the Quantitative Trait Loci Mapping Using Crossbred Population2005 December;18(12)





 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print




