 |
 |
Abstract
Fat mass and obesity associated gene (FTO) plays an important role in appetite control and energy consumption in human and mice. In order to examine FTO expression influence on fat deposition in Suzhong pigs, FTO mRNA expression was detected in 16 tissues by RT-PCR, FTO protein expression was detected in 5 tissues by western blot, and association of FTO polymorphism with meat quality traits was analyzed in Suzhong populations with 714 records. RT-PCR results revealed that FTO mRNA was expressed in all sixteen tissues with significant differences (p<0.05), expression in backfat was significantly higher than that of any other tissue (p<0.05), and expression in longissimus dorsi muscle had the second highest significance level (p<0.05). Western blot results demonstrated that FTO protein was highly expressed in backfat and longissimus dorsi muscle. Furthermore, FTO mRNA and protein expression in tissues of high-fat pigs was significantly higher than that of low-fat pigs (p<0.05), suggesting FTO expression had advantageous effects on fat deposition. FTO polymorphism results evidenced that at A227G locus, G allele seemed to have advantageous effects on fat deposition, indicating it could be a significant candidate gene for improving pork quality in Suzhong pigs.
Pork quality is an increasing concern for swine producers due to the development of export markets and increased consumer demands (Van Wijk et al., 2005; 2006). Fat deposition, whether in adipose tissue or muscle, contributes importantly to various aspects of meat quality and are central to the nutritional value of meat (Wood et al., 2008), and fat deposition could influence some economic meat quality traits, such as backfat thickness, Intramuscular fat (IMF) content and lean meat percentage. It was reported that each type of fat deposition can have a different developmental profile (Kouba et al., 1999; Hausman and Poulos, 2004). Additionally, obesity in humans is an increasing problem worldwide, and research on candidate genes in good animal models is urgently required. The pig is an excellent model as its metabolism, organ size, and eating habits resemble humans (Madsen et al., 2009). Therefore, for these reasons itŌĆÖs urgent to study the molecular mechanism of fat deposition in pigs.
Many major genes affecting meat quality traits in pigs have been successfully identified with the development of candidate gene and comparative mapping approaches, such as fatty acid-binding protein (FABP) (Gerbens et al., 2000) and proliferator-activated receptors (PPARs) (Mandard et al., 2004), Lipoprotein lipase (LPL) (Shen et al., 2010). Fat mass and obesity associated gene (FTO) was discovered in 2007, and FTO SNPs were associated with obesity of children and adults. A genome-wide study found that the FTO gene was located on chromosome 6 (Du et al., 2008). The region on chromosome 6 was first indicated as the site of the FTO gene as quantitative trait loci (QTLs) linked to regulation of some fat traits in pigs were mapped to the region (Hu and Reecy, 2007). In addition, swine FTO polymorphism was associated with pork quality, such as intramuscular fat deposition, analysis (Dina et al., 2007; Frayling et al., 2007). In humans and mice, some studies reported that FTO played an important role in appetite control and energy consumption (Kl├Čting et al., 2008; Fischer et al., 2009; Olszewski et al., 2009).
Suzhong pig is a new breed with excellent reproductive performance and meat quality originating from Taihu pig. The objective of this study is to examine the role of FTO expression on regulation of fat deposition in Suzhong pigs. To achieve this target, we cloned the coding sequence (CDS) of swine FTO gene in Suzhong pigs, analyzed the FTO mRNA expression by RT-PCR, detected the FTO protein expression by western blot, and examined the association of FTO polymorphism with meat quality traits.
Animals were all healthy half-siblings fed under the same conditions from the seed stock farms of Hongze Xinxiang Swine Breeding Company (Jiangsu, China). In order to investigate FTO expression, ten Suzhong pigs (five high-fat pigs and five low-fat pigs) were slaughtered on the same day by electrical stunning when their body-weight was 95┬▒2 kg at six-month age. Afterwards, 16 tissues of heart, liver, kidney, longissimus dorsi muscle, backfat, lung, stomach, bladder, spleen, uterus, cervix, ovary, large intestine, oviduct, endometrium and small intestine were collected and prepared according to the procedure proposed by Lord with minor modifications (Lord et al., 2006). For RT-PCR and western blot, these tissues were immediately saved into liquid nitrogen. For meat quality traits, longissimus dorsi muscle area was tested using a planimeter (Haguang, China), and lean meat percentage was tested using a lean meat percentage analyzer (SFK, Denmark) (Table 1).
To detect FTO polymorphism, there were 714 records of 51 pigs killed, these records contained such data as: ear number, birth date, breed, parity, longissimus dorsi muscle area and lean meat percentage. Ear tissues of pigs were collected in centrifuge tubes (1.5 mL) with 70% ethanol, stored at 4┬░C until the genomic DNA of each ear sample was extracted using a Genomic DNA Rapid Isolation Kit (Sangon, China) (Fu et al., 2012b).
Primers for swine FTO (GenBank: NM_001112692) and housekeeping gene (GAPDH) were designed by Oligo 6 software and synthesized by Introvigen company (Shanghai, China). Primer FTO_1 and FTO_2 was used in RT-PCR, and Primer FTO_3 was used in detection of FTO polymorphism. Both Primer FTO_1 and FTO_2 were designed as cross-exons primers (Table 2).
Total RNA was extracted from swine tissues using a modified Trizol/chloroform (Invitrogen, USA) extraction according to the protocol (Sambrook and Russell, 2001). RNA samples were quantified spectrophotometrically (Shimadzu, Japan) and the integrity was confirmed using 1.5% agarose gel.
In reverse transcriptase-polymerase chain reaction (RT-PCR), total RNA was reverse transcribed using a First Strand cDNA Synthesis Kit (Fermentas, Canada) and was carried out in a 25 ╬╝L reaction volume by two steps according to the kit manual. The amplifying reactions were carried out in a 15 ╬╝L reaction volume containing 1 ╬╝L cDNA, 0.2 mmol/L dNTPs, 1.5 mmol/L MgCl2, 0.5 ╬╝M of each PCR primer, 1 U Taq DNA polymerase and reaction buffer (Takara, Japan). The amplifying conditions were heating at 94┬░C for 5 min, followed by 35 cycles of denaturation at 94┬░C for 30 s, annealing at 60┬░C for 30 s, extension at 72┬░C for 50 s, after that, extension at 72┬░C for 7 min. Amplification of the house-keeping gene GAPDH was performed asthe control for the reaction. The PCR products were confirmed by electrophoresis on 1.5% agarose gel.
The Real-time PCR amplification was performed in a 20 ╬╝L reaction volume, containing 10 ╬╝L 2├Ś Real-time PCR Master Mix (SYBR Green), 1 ╬╝L cDNA, 1 ╬╝L of each PCR primer (10 ╬╝M), 1 ╬╝L Taq DNA polymerase (10 u/╬╝L) and 6 ╬╝L sterilized water (Toyobo, Japan). The reaction was run on a Line-gene Fluorescence PCR detection system (Bori, China) heating at 95┬░C for 5 min for initial enzyme activation, followed by 45 cycles amplification including denaturation at 95┬░C for 20 s, annealing at 60┬░C for 25 s, and extension at 72┬░C for 30 s. Adequate precautions were given to prevent cross-contamination and negative controls were performed routinely with each reaction. The standard curves were prepared for both tested and housekeeping gene, GAPDH (GenBank: U48832).
Frozen sections of endometrial specimens were prepared and western-blot was performed as previously described (Wong and Medrano, 2005) with minor modification. Tissues were cracked in tissue protein extracts (0.05 mol/L Tris┬ĘHCl, NaCl 8.76 mg/mL, 1% TritonX-100 and 100 ug/mL PMSF) (Keygen, China) by vortex meter (Huxi, China). Total protein concentrations were detected using the BCA Protein Assay Kit (Keygen, China) according to the manufacturerŌĆÖs recommendations.
There was a 60 ╬╝g sample separated from a 10% Tris┬ĘHCl polyacrylamide gel using electrophoresis system (Bio-Rad, America), and protein from the gel was transferred onto a single PVDF membrane (Bio-Rad, America). After rinsing in TBST for 5 min, the membrane was soaked in 5% skim milk (in TBST) for 1 h. Next, the membrane was immerged into 1:200 dilution of the the primary antibody (mouse anti-human FTO mAb, Santa Cruz, USA) at 4┬░C for 1 h. After rinsing in TBST for 5 min, the membrane was immerged into 1:5,000 dilution of the secondary antibody (HRP) (Santa Cruz, USA) for 2 h. Finally, the membrane was colored using the DAB kit (Invitrogen, USA) and exposed using Chemiluminescence Detection Kit for HRP (Keygen, China). ╬▓-Actin was used as a housekeeping gene. Scanned images were quantified using LabWorks gray image analysis software (Fu et al., 2012a).
For purpose of detecting single nucleotide polymorphisms in the coding sequence (cSNPs), a 609-bp cDNA sequence was scanned within Exon 3 of the swine FTO gene by PCR-sequencing technology. The amplification reactions were performed in a 15 ╬╝L reaction volume containing 50 ng genomic DNA, 1.5 ╬╝L of 10├ŚPCR buffer (containing 100 mM Tris-HCl (pH 8.0), 500 mM KCl, 10 mM of MgCl2 and 0.1% Triton X-100), 200 ╬╝M of each dNTPs, 0.5 ╬╝L of each primer (10 pM) and 3 U of Taq DNA polymerase (Takara, China). The PCR conditions were heating at 94┬░C for 5 min, followed by 35 cycles of reactions containing denaturation at 94┬░C for 30 s, annealing at 59┬░C for 30 s, and extension at 72┬░C for 60 s, finally, a further heating at 72┬░C for 7 min. The PCR products were confirmed by electrophoresis on 1.5% agarose gel and then purified using a DNA fragment gel purification and extraction kit (Sangon, China).
Direct DNA sequencing was implemented using purified PCR products on an ABI 3730XL automated DNA sequencer (Applied Biosystems, USA) by a professional gene sequencing company (Invitrogen, Shanghai, China). Each sample was sequenced twice and all bases of both strands were sequenced. The sequence data were analyzed using DNAMAN (Version 5.2) and aligned with the consensus sequence of swine FTO sequence available in the GenBank database.
Data analysis was performed using GLM procedures of SAS (Ver. 8.2). Significant differences were established utilizing PDIFF option of GLM with least significant differences (LSD) post hoc test and assumed as statistically significant for pŌēż0.05, and Lsmeans┬▒SE are presented. The additive and dominance effects were estimated using Estimate step of GLM procedure in SAS according to the expressions a = (AAŌłÆBB)/2 and d = ABŌłÆ(AA+BB)/2, respectively, where AA, AB, and BB are the genotypic values (Tang et al., 2010; Fu et al., 2012b).
RT-PCR data were examined by gray values ratio (FTO/GAPDH) of scanned agarose lane images by LabImage software, real-Time PCR data were analyzed using delta-delta CT method (also known as the 2ŌłÆ╬ö╬öCT method) (Livak and Schmittgen, 2001; Schmittgen and Livak, 2008), GAPDH gene is as positive control.
Association analysis between FTO polymorphism and meat quality traits were evaluated with the GLM procedure of SAS (Version 8.2). Genotype effects of SNPs were analyzed by the established model (Fu et al., 2012b),
Where Y is the meat quality trait, ╬╝ is the overall mean, HYSi is the effect of herd-year-season (i = 1), Bj is the effect of breed (j = 1), Sk is the effect of sex (k = 1, 2), Gl is the effect of genotype (l = 1, 2, 3) and e is the random residual.
According to OD values measured by spectrophotometer, the OD260/OD280 values of total RNA extracted from swine tissues ranged from 1.8 to 2.1, and RNA content ranged from 0.60 to 2.22 ╬╝g/╬╝L, suggesting the quality of RNA extracted was good and it could be used for subsequent experiments.
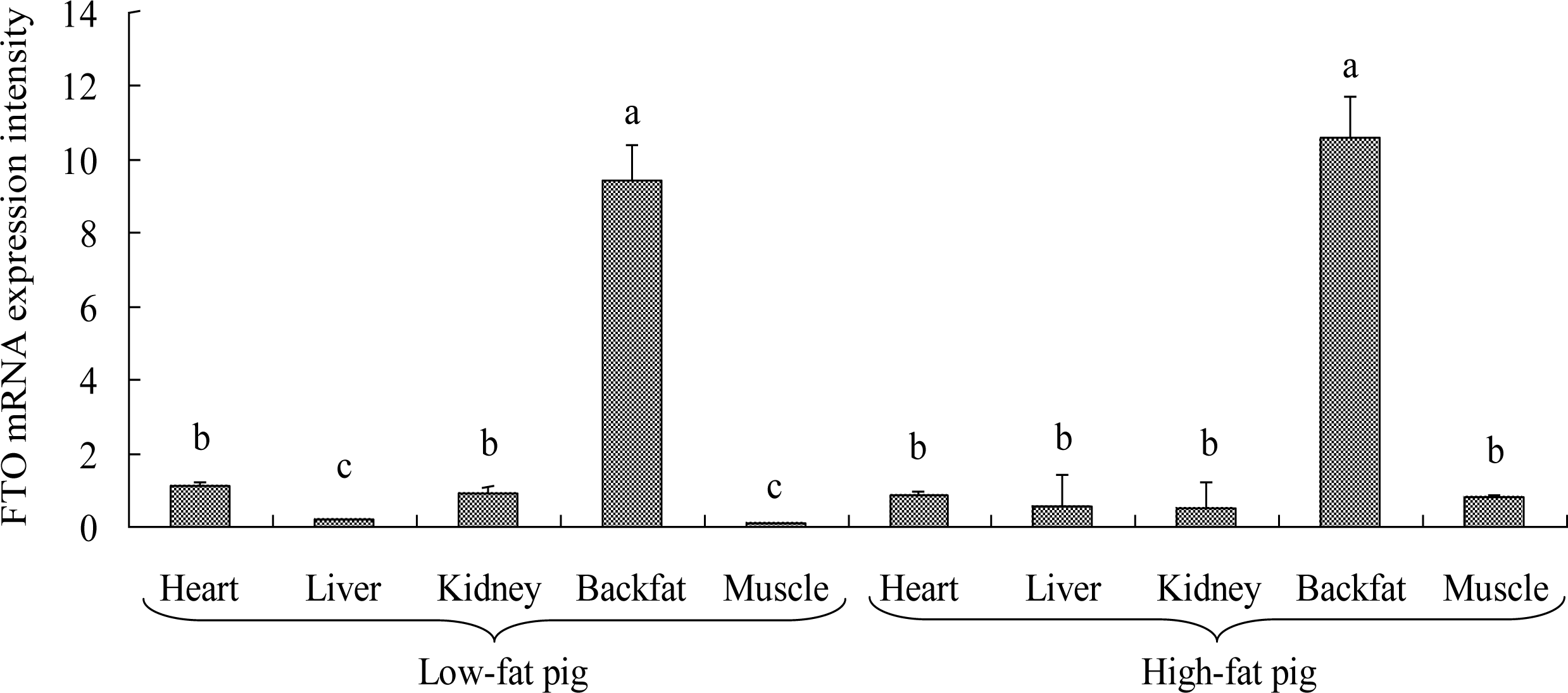
Expression analysis showed that FTO mRNA was expressed in 16 tissues in Suzhong pigs. Ranking by mRNA expression intensity (FTO/GAPDH) in these tissues, from highest to lowest, it was backfat (9.44), heart (1.10), kidney (0.92), ovary (0.91), lung (0.89), oviduct (0.86), endometrium (0.83), longissimus dorsi muscle (0.81), large intestine (0.66), uterus (0.65), cervix (0.64), spleen (0.57), bladder (0.50), stomach (0.42), liver (0.20) and small intestine (0.04). Among all these tissues, mRNA expression in backfat was extremely significantly (p<0.01) higher than any other tissue and expression in longissimus dorsi muscle of high-fat pigs was significantly (p<0.05) higher than that of low-fat pigs (Figures 1 and 2).
This suggested that a higher FTO expression was in favor of a higher fat content, a lower longissimus dorsi muscle area and a lower lean meat percentage (Table 1), FTO expression seemed to have advantageous effects on fat deposition.
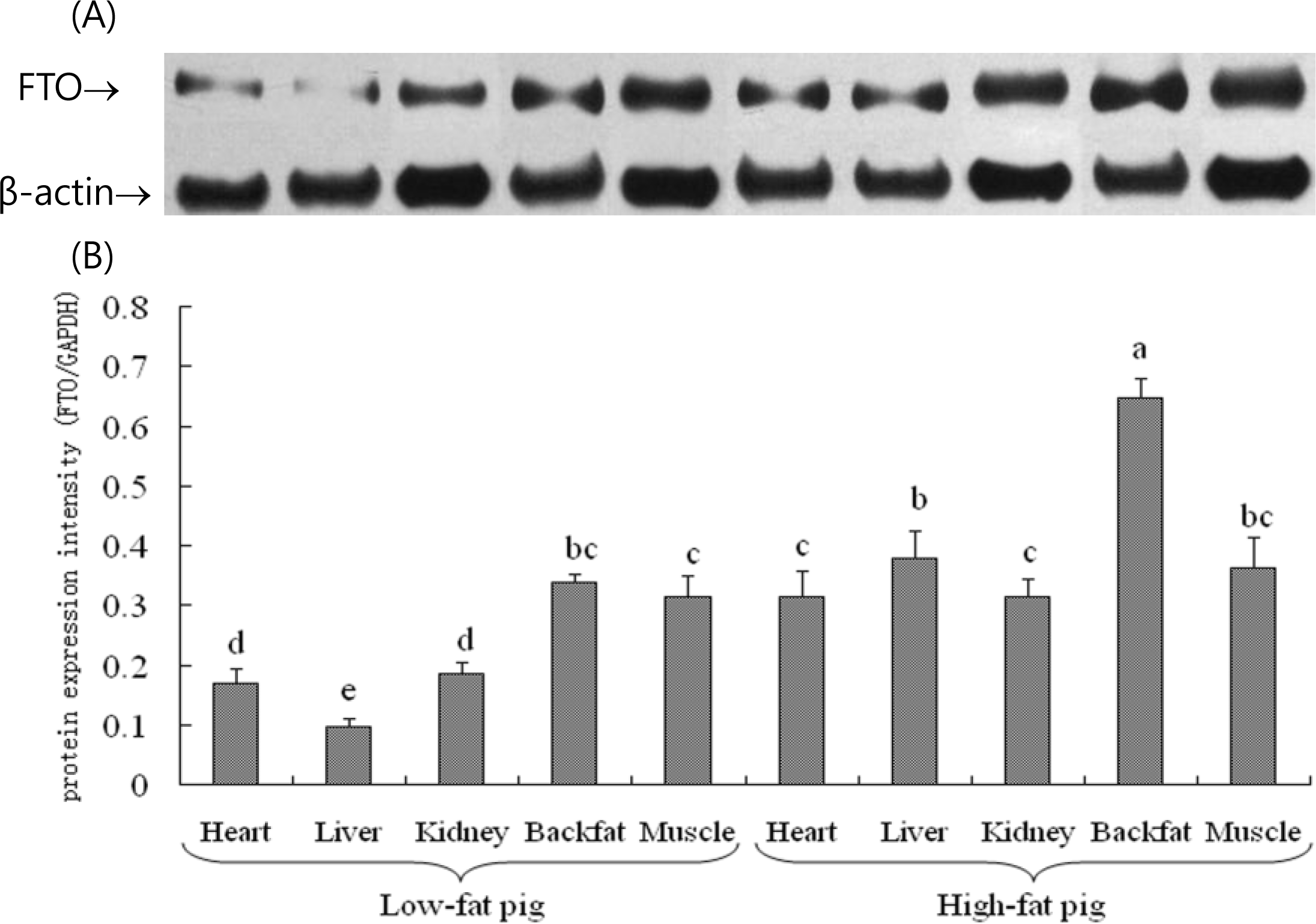
The western blot results showed that FTO protein (GenBank: NP_001106162) was expressed in heart, liver, kidney, longissimus dorsi muscle and backfat of Suzhong pigs. Ranking by FTO protein expression intensity, from highest to lowest, it was backfat, longissimus dorsi muscle, kidney, heart, liver in low-fat pigs, and itŌĆÖs backfat, longissimus dorsi muscle, liver, kidney, heart in high-fat pigs. FTO protein expression in backfat was significantly (p<0.05) higher than that of any other tissue in all pigs, and FTO protein expression in longissimus dorsi muscle was second highest (Figure 3).
FTO protein expression in tissues of high-fat pigs was extremely significantly (p<0.01) higher than that of low-fat pigs, and the expression difference between high-fat pigs and low-fat pigs was consistent with the real-time PCR results (Table 3). Considering that longissimus dorsi muscle area and lean meat percentage of high-fat pigs were both significantly (p<0.05) lower than that of low-fat pigs (Table 1), the results showed that a higher FTO expression might conducive to a lower longissimus dorsi muscle area and lean meat percentage, which was opposite to fat content. Consequently, FTO might be involved in regulating the swine fat deposition.
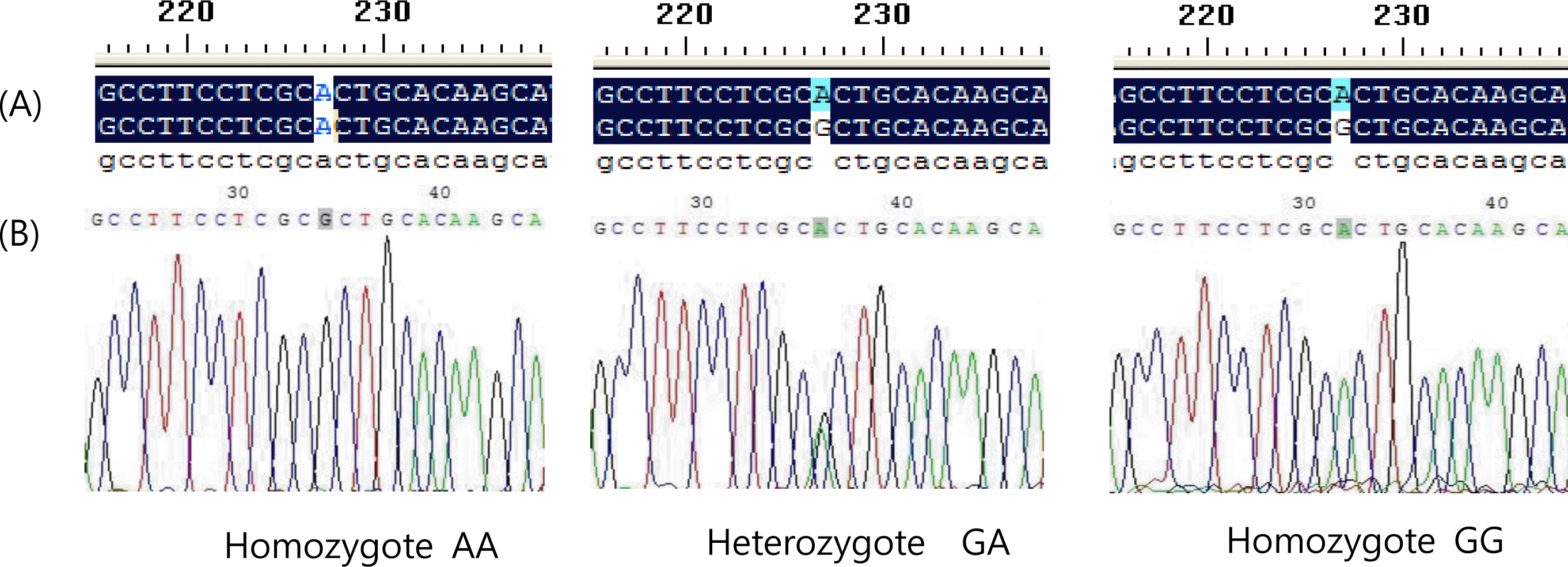
The coding sequence (CDS, 15 to 1,532 bp long) of swine FTO was cloned from Suzhong pigs (GenBank: JX873956), and there were 10 SNPs discovered, including G52A, A227G, C523T, C608G, G628A, A655G, A810G, A886G, G1002A and A1344G. Among these SNPs, the A227G (at position 227 bp of cDNA sequence, an A/G transition) within Exon 3 (609-bp long) of swine FTO was polymorphic in 51 Suzhong pigs. At A227G locus, three PCR products of each genotype were random selected and sequenced by automated sequencing to validate the results. If position 227 is A, then it was designated as the A allele, G for the G allele (Figures 4 and 5).
All genotypes of FTO occurred in the Suzhong pigs, which were found to be in Hardy-Weinberg equilibrium for the genotyped locus through the chi-square test (p>0.05). The allele and genotype frequencies are shown in Table 4.
The data of meat quality traits were analyzed by genotype, and these traits included longissimus dorsi muscle area/weight, lean meat percentage, PH value, luminosity, ŌĆ£aŌĆØ, ŌĆ£bŌĆØ, meat color score, marbling score and water holding capacity. For longissimus dorsi muscle area/weight, lean meat percentage, PH value, meat color score and water holding capacity, those of Genotype AA were significantly (p<0.05) higher than those of Genotype GG.In contrast, for luminosity and marbling score, those of Genotype AA were lower than those of Genotype GG significantly (p<0.05). These results suggested that G allele seemed to have advantageous effects on fat deposition (Table 5).
Fat deposition could influence meat-quality traits, such as backfat thickness, longissimus dorsi muscle area, intramuscular fat (IMF) content and lean meat percentage, etc (Wood et al., 2008). These meat quality traits were all important breeding goals for pig producers, because they are closely related to pork quality, which is positively correlated with meat tenderness, juiciness, and taste (DeVol et al., 1988; Cui et al., 2011). Recently, there have been some genes reported to be associated with fat deposition, such as Apolipoprotein M (APOM) (Pan et al., 2010), diacylgycerol acyltransferase (DGAT) (Cui et al., 2011), nudix (nucleoside diphosphate linked moiety X)-type motif 6 (Nudt6) (Sun et al., 2012) as well as others. Fat mass and obesity associated gene (FTO) played an important role in appetite control and energy consumption in human and mice (Kl├Čting et al., 2008; Fischer et al., 2009; Olszewski et al., 2009), therefore the present research studied the FTO mRNA expression in 16 tissues and protein expression in 5 tissues, examined FTO expression difference between low-fat pigs and high-fat pigs, and analyzed association of FTO polymorphism with meat quality traits in a local pig breed of China.
At the RNA level, thereŌĆÖre 16 tissues of FTO mRNA expression in Suzhong pigs with significant differences (p<0.05) by RT-PCR, ranking by mRNA expression intensity from highest to lowest, it was backfat, heart, kidney, ovary, lung, oviduct, endometrium, longissimus dorsi muscle, large intestine, uterus, cervix, spleen, bladder, stomach, liver and small intestine in turn. In G├Čttingen minipigs, FTO mRNA expression was detected in muscle, brain, cerebellum, hippocampus, liver, kidney and heart with significantly higher levels in brain tissues (p<0.05) (Madsen et al., 2009). In sheep, FTO mRNA expression was detected in hypothalamus, pancreas, heart, kidney, adipose tissues and liver and skeletal muscle (S├®bert et al., 2010). At protein level, western blot results indicated that FTO protein was detected in all five tissues of Suzhong pigs with a significantly higher expression in backfat than that of any other tissue (p<0.05)). Differences between mRNA and protein expression might be due to different regulation of these genes on translation level (Fu et al., 2012a).
Both FTO mRNA and protein expression in tissues of high-fat pigs was higher than that of low-fat pigs at significant levels (p<0.05), and lean meat percentage of high-fat pigs was significantly lower than that of low-fat pigs (p<0.05), this might suggest that FTO expression was inversely proportional to lean meat percentage, and lean meat percentage was inversely proportional to fat content, so FTO expression had advantageous effects on fat deposition. Related studies have reported that obese animals exhibited a significant upregulation of FTO mRNA abundance (S├®bert et al., 2010), while in humans, allelic distributions of FTO SNPs were similarly related to alterations of energy intake in childhood (Timpson et al., 2008; Wardle et al., 2009).
At the A227G locus, the meat quality traits of longissimus dorsi muscle area/weight, lean meat percentage, PH value, meat color score and water holding capacity were significantly higher in pigs with Genotype AA than those of genotype GG (p<0.05). In contrast, for meat quality traits of luminosity and marbling score, were significantly (p<0.05) lower in Genotype AA than those of Genotype GG, suggesting G allele seemed to have advantageous effects on fat deposition. In Italian Duroc pigs, FTO polymorphism was associated with fat deposition traits (Fontanesi et al., 2009). In an Italian Duroc population and a Berkshire├ŚYorkshire population, two SNPs were detected in Exon 3 and Intron 4 of swine FTO, and the mutation effects on obesity-related traits were also confirmed (Fontanesi et al., 2010). The cSNP in Exon 3 of FTO did not directly alter the amino acid residue, which indicates that it was possible the polymorphism indirectly affects reproductive traits by being in linkage disequilibrium with another polymorphism that directly influences the analyzed quantitative traits (Niu et al., 2009; Fu et al., 2012b).
In conclusion, the study showed both FTO mRNA and protein expression in tissues of high-fat pigs was significantly higher than that of low-fat pigs, and at A227G locus, the G allele seemed to have advantageous effects on fat deposition. This suggested that FTO might be involved in fat deposition mechanisms, and would be one of the major genes affecting meat quality traits.
ACKNOWLEDGMENTS
This research was supported by National Natural Science Foundation of China (No. 31201767), Postdoctoral Research Funding Project of Jiangsu Province (No. 026056511104), Jiangsu Agricultural Sciences and Technology Autonomy Innovation Fund Project (NO. CX(13)5042) and Nanjing Station Foundation of National Modern Swine Industrial-Technology System (No. nycytx-009). These authors greatly appreciate Hongze Xinxiang Swine Breeding Company (Jiangsu, China) for assistance in collecting experimental materials.
Figure┬Ā1.
The 1.5% agarose gel electrophoresis atlas for the expression of swine fat mass and obesity associated gene (FTO) in different tissues of Suzhong pigs, and gray values of scanned agarose lane images by LabImage software, GAPDH gene is as positive control. (A) agarose gel electrophoresis atlas, (B) Gray values of FTO/GAPDH. Values with different superscripts show significant levels within columns (p<0.05).
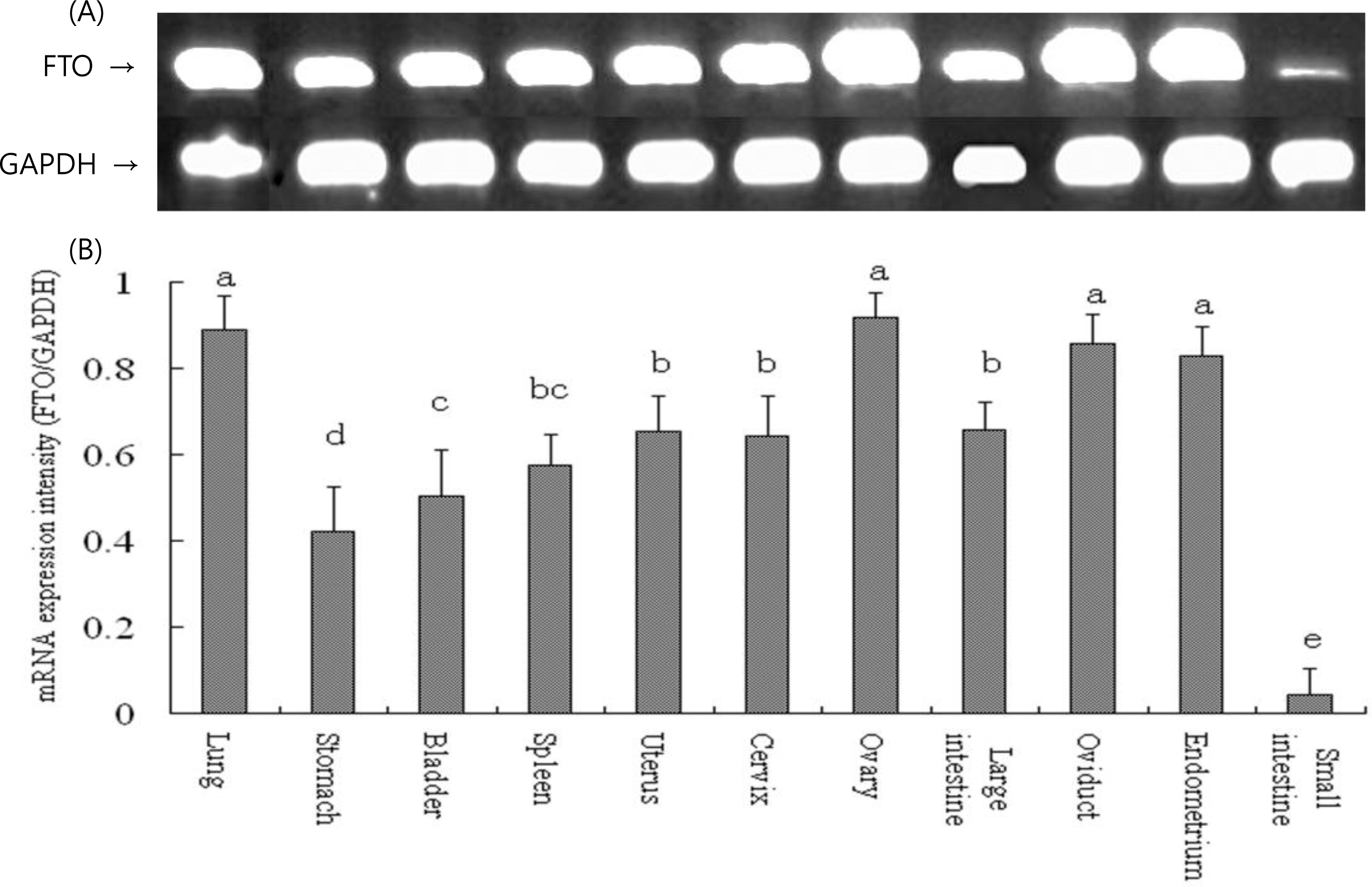
Figure┬Ā2.
The mRNA expression intensity of fat mass and obesity associated gene (FTO) in different tissues. Real-time PCR detected mRNA expression in heart, liver, kidney, backfat and longissimus dorsi muscle (muscle) of Suzhong pigs. GAPDH was used as the housekeeping gene. The mRNA expression intensity of FTO varied significantly in different tissues.
Figure┬Ā3.
Western-blot results of FTO expression in different tissues of Suzhong pigs, including heart, liver, kidney, backfat and longissimus dorsi muscle (muscle). (A) Western blot lanes (B) Gray values of FTO/╬▓-actin for scanned western blot lane images. Lowercase letters on columns indicate significant levels within columns (p<0.05).
Figure┬Ā4.
The PCR-Sequencing results for the FTO gene polymorphism, (A) comparison results using DNAMAN software, (B) Peak value figure.
Figure┬Ā5.
The coding sequence (CDS, 15-1,532 bp) of fat mass and obesity associated gene (FTO) in Suzhong pigs by cloning and sequencing, and a new GenBank accession number was obtained (GenBank: JX873956), sequence with a gray color background was the cloning sequence in Suzhong pigs.

Table┬Ā1.
Meat quality traits related with fat deposition in Suzhong pigs for FTO expression trial (Mean┬▒SE)
| Item | Longissimus dorsi muscle area | Lean meat percentage |
Backfat thickness
|
Marbling score | ||
|---|---|---|---|---|---|---|
| Shoulder at the thickest joint | Thoracic and lumbar spine junction | Lumbar and sacral spine junction | ||||
| Low-fat pigs | 61.86┬▒2.46a | 56.62┬▒1.35a | 3.9┬▒0.53a | 2.4┬▒0.46 | 2.1┬▒0.48a | 4.71┬▒1.12 |
| High-fat pigs | 48.52┬▒3.57b | 49.42┬▒2.39b | 5.2┬▒1.21b | 2.7┬▒0.56 | 3.2┬▒0.8b | 4.72┬▒1.33 |
Table┬Ā2.
Primer pairs and PCR conditions used in RT-PCR and SNPs detection analysis of FTO
Table┬Ā3.
Change in protein expression intensity of fat mass and obesity associated gene (FTO) in different tissues (Mean┬▒SE)
| Item | FTO/╬▓-actin | |
|---|---|---|
| Tissue | Heart | 0.24┬▒0.03Bc |
| Liver | 0.24┬▒0.04Bc | |
| Kidney | 0.49┬▒0.05Aa | |
| Longissimus dorsi muscle | 0.34┬▒0.03Bb | |
| Backfat | 0.25┬▒0.03Bc | |
| Item | Low-fat pig | 0.22┬▒0.02A |
| High-fat pig | 0.40┬▒0.03B |
Table┬Ā4.
Allele and genotype frequencies of FTO in Suzhong pigs
| NO. of pigs |
Genotype distribution
|
Allele frequencies
|
x2 | |||
|---|---|---|---|---|---|---|
| AA | GA | GG | C | T | ||
| 51 | 30 | 18 | 3 | 0.76 | 0.24 | 0.01 |
Table┬Ā5.
Effects of FTO genotype on meat quality traits (LS Means┬▒SE)
| Meat quality traits |
Genotype
|
Additive effect | Dominant effect | ||
|---|---|---|---|---|---|
| AA | GA | GG | |||
| Longissimus dorsi muscle area/weight | 0.57┬▒0.01Aa | 0.55┬▒0.02Aa | 0.37┬▒0.04Bb | 1.00┬▒0.02 | 0.08┬▒0.03 |
| Lean meat percentage | 55.78┬▒0.98A | 51.34┬▒1.25B | 40.54┬▒3.06C | 7.62┬▒1.61 | 3.18┬▒2.03 |
| PH value | 5.83┬▒0.05ab | 5.91┬▒0.06a | 5.57┬▒0.16b | 0.13┬▒0.08 | 0.21┬▒0.11 |
| Luminosity (L) | 41.09┬▒1.04Aa | 41.54┬▒1.32Aa | 46.50┬▒3.22Ab | ŌłÆ2.71┬▒1.70 | ŌłÆ2.26┬▒2.14 |
| a (range from magenta to green) | 6.35┬▒0.46 | 5.29┬▒0.58 | 6.15┬▒1.42 | 0.09┬▒0.75 | ŌłÆ0.97┬▒0.94 |
| b (range from yellow to blue) | 3.50┬▒0.29 | 3.07┬▒0.37 | 3.38┬▒0.90 | 0.06┬▒0.48 | ŌłÆ0.37┬▒0.60 |
| Meat color score | 3.55┬▒0.11Aa | 3.54┬▒0.14Aa | 2.69┬▒0.35Ab | 0.43┬▒0.18 | 0.42┬▒0.23 |
| Marbling score | 4.46┬▒0.22Aa | 4.91┬▒0.28Aab | 6.03┬▒0.69Ab | ŌłÆ0.78┬▒0.37 | ŌłÆ0.34┬▒0.46 |
| Water holding capacity | 16.69┬▒0.77Aa | 13.60┬▒0.98 Ab | 14.11┬▒2.38Aab | 1.29┬▒1.26 | ŌłÆ1.80┬▒1.58 |
Longissimus dorsi muscle area/weight was values of longissimus dorsi muscle area divided by values of weight in pigs. Luminosity (L), a and b were three parameters of meat color measured using chroma meter cr400 (Konica Minolta, Japan). Among these parameters, values of L ranged from 0 to 100 (color range from black to white), values of a ranged from 127 to ŌłÆ128 (color range from magenta to green) and values of b ranged from 127 to ŌłÆ128 (color range from yellow to blue). Values of meat color score ranged from 1.0 to 6.0 (color range from white to red), and values of marbling score ranged from 1.0 to 10.0, both of them were measured using American shade guide. Values with different superscripts show significant levels within rows
REFERENCES
Cui J, Zeng Y, Wang H, Chen W, Du J, Chen Q, Hu Y, Yang L. 2011. The effects of DGAT1 and DGAT2 mRNA expression on fat deposition in fatty and lean breeds of pig. Livest Sci 140:292ŌĆō296.

DeVol D, McKeith F, Bechtel PJ, Novakofski J, Shanks R, Carr T. 1988. Variation in composition and palatability traits and relationships between muscle characteristics and palatability in a random sample of pork carcasses. J Anim Sci 66:385ŌĆō395.

Dina C, Meyre D, Gallina S, Durand E, K├Črner A, Jacobson P, Carlsson LMS, Kiess W, Vatin V, Lecoeur C. 2007. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet 39:724ŌĆō726.


Du ZQ, Fan B, Zhao X, Amoako R, Rothschild MF. 2008. Association analyses between type 2 diabetes genes and obesity traits in pigs. Obesity 17:323ŌĆō329.


Fischer J, Koch L, Emmerling C, Vierkotten J, Peters T, Br├╝ning JC, R├╝ther U. 2009. Inactivation of the Fto gene protects from obesity. Nature 458:894ŌĆō898.


Fontanesi L, Scotti E, Buttazzoni L, DallŌĆÖOlio S, Bagnato A, Lo Fiego DP, Davoli R, Russo V. 2010. Confirmed association between a single nucleotide polymorphism in the FTO gene and obesity-related traits in heavy pigs. Mol Biol Rep 37:461ŌĆō466.


Fontanesi L, Scotti E, Buttazzoni L, Davoli R, Russo V. 2009. The porcine fat mass and obesity associated (FTO) gene is associated with fat deposition in Italian Duroc pigs. Anim Genet 40:90ŌĆō93.


Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JRB, Elliott KS, Lango H, Rayner NW. 2007. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316:889ŌĆō894.



Fu Y, Fu J, Ren Q, Chen X, Wang A. 2012a. Expression of Eph A molecules during swine embryo implantation. Mol Biol Rep 39:2179ŌĆō2185.


Fu Y, Fu J, Wang A. 2012b. Association of EphA4 polymorphism with swine reproductive traits and mRNA expression of EphA4 during embryo implantation. Mol Biol Rep 39:2689ŌĆō2696.


Gerbens F, De Koning D, Harders F, Meuwissen T, Janss L, Groenen M, Veerkamp J, Van Arendonk J, Te Pas M. 2000. The effect of adipocyte and heart fatty acid-binding protein genes on intramuscular fat and backfat content in Meishan crossbred pigs. J Anim Sci 78:552ŌĆō559.


Hausman G, Poulos S. 2004. Recruitment and differentiation of intramuscular preadipocytes in stromal-vascular cell cultures derived from neonatal pig semitendinosus muscles. J Anim Sci 82:429ŌĆō437.


Kl├Čting N, Schleinitz D, Ruschke K, Berndt J, Fasshauer M, T├Čnjes A, Sch├Čn M, Kovacs P, Stumvoll M, Bl├╝her M. 2008. Inverse relationship between obesity and FTO gene expression in visceral adipose tissue in humans. Diabetologia 51:641ŌĆō647.


Kouba M, Bonneau M, Noblet J. 1999. Relative development of subcutaneous, intermuscular, and kidney fat in growing pigs with different body compositions. J Anim Sci 77:622ŌĆō629.


Livak K, Schmittgen T. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-[Delta][Delta] CT method. Methods 25:402ŌĆō408.


Lord E, Murphy B, Desmarais J, Ledoux S, Beaudry D, Palin M. 2006. Modulation of peroxisome proliferator-activated receptor ╬┤ and ╬│ transcripts in swine endometrial tissue during early gestation. Reproduction 131:929ŌĆō942.


Madsen MB, Birck MM, Fredholm M, Cirera S. 2009. Expression studies of the obesity candidate gene FTO in pig. Anim Biotechnol 21:51ŌĆō63.

Mandard S, M├╝ller M, Kersten S. 2004. Peroxisome proliferator-activated receptor a target genes. Cell Mol Life Sci 61:393ŌĆō416.


Niu B, Ye L, Li F, Deng C, Jiang S, Lei M, Xiong Y. 2009. Identification of polymorphism and association analysis with reproductive traits in the porcine RNF4 gene. Anim Reprod Sci 110:283ŌĆō292.


Olszewski P, Fredriksson R, Olszewska A, Stephansson O, Alsi├Č J, Radomska K, Levine A, Schi├Čth H. 2009. Hypothalamic FTO is associated with the regulation of energy intake not feeding reward. BMC Neurosci 10:129ŌĆō140.



Pan G, Fu Y, Zuo B, Ren Z, Xu D, Lei M, Zheng R, Xiong YZ. 2010. Molecular characterization, expression profile and association analysis with fat deposition traits of the porcine APOM gene. Mol Biol Rep 37:1363ŌĆō1371.


S├®bert S, Hyatt M, Chan L, Yiallourides M, Fainberg H, Patel N, Sharkey D, Stephenson T, Rhind S, Bell R. 2010. Influence of prenatal nutrition and obesity on tissue specific fat mass and obesity-associated (FTO) gene expression. Reproduction 139:265ŌĆō274.


Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. Cold spring harbor laboratory press.
Schmittgen T, Livak K. 2008. Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101ŌĆō1108.


Shen Y, Lookene A, Zhang L, Olivecrona G. 2010. Site-directed mutagenesis of apolipoprotein CII to probe the role of its secondary structure for activation of lipoprotein lipase. J Biol Chem 285:7484ŌĆō7492.



Sun L, Yu S, Wang H, Fan B, Liu B. 2012. NUDT6, the FGF-2ŌĆÖs antisense gene, showed associations with fat deposition related traits in pigs. Mol Biol Rep 39:4119ŌĆō4126.


Tang S, Sun D, Ou J, Zhang Y, Xu G. 2010. Evaluation of the IGFs (IGF1 and IGF2) genes as candidates for growth, body measurement, carcass, and reproduction traits in Beijing You and Silkie chickens. Anim Biotechnol 21:104ŌĆō113.


Timpson NJ, Emmett PM, Frayling TM, Rogers I, Hattersley AT, McCarthy MI, Davey Smith G. 2008. The fat massŌĆōand obesity-associated locus and dietary intake in children. Am J Clin Nutr 88:971ŌĆō978.



Van Wijk H, Arts D, Matthews J, Webster M, Ducro B, Knol E. 2005. Genetic parameters for carcass composition and pork quality estimated in a commercial production chain. J Anim Sci 83:324ŌĆō333.


Van Wijk H, Dibbits B, Baron E, Brings A, Harlizius B, Groenen M, Knol E, Bovenhuis H. 2006. Identification of quantitative trait loci for carcass composition and pork quality traits in a commercial finishing cross. J Anim Sci 84:789ŌĆō799.







 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print





