 |
 |
| Anim Biosci > Volume 35(3); 2022 > Article |
|
Abstract
Objective
The highly pathogenic avian influenza virus (HPAIV) is a threat to the poultry industry as well as the economy and remains a potential source of pandemic infection in humans. Antiviral genes are considered a potential factor for HPAIV resistance. Therefore, in this study, we investigated gene expression related to cytokine-cytokine receptor interactions by comparing resistant and susceptible Ri chicken lines for avian influenza virus infection.
Methods
Ri chickens of resistant (Mx/A; BF2/B21) and susceptible (Mx/G; BF2/B13) lines were selected by genotyping the Mx dynamin like GTPase (Mx) and major histocompatibility complex class I antigen BF2 genes. These chickens were then infected with influenza A virus subtype H5N1, and their lung tissues were collected for RNA sequencing.
Results
In total, 972 differentially expressed genes (DEGs) were observed between resistant and susceptible Ri chickens, according to the gene ontology and Kyoto encyclopedia of genes and genomes pathways. In particular, DEGs associated with cytokine-cytokine receptor interactions were most abundant. The expression levels of cytokines (interleukin-1β [IL-1β], IL-6, IL-8, and IL-18), chemokines (C-C Motif chemokine ligand 4 [CCL4] and CCL17), interferons (IFN-γ), and IFN-stimulated genes (Mx1, CCL19, 2′-5′-oligoadenylate synthase-like, and protein kinase R) were higher in H5N1-resistant chickens than in H5N1-susceptible chickens.
Avian influenza (AI) is a virus of the genus influenzavirus A in the Orthomyxoviridae family of viruses [1]. Based on genetics and disease severity, AI viruses are classified either as highly pathogenic avian influenza (HPAI) or lowly pathogenic AI [2]. In particular, a high number of deaths were observed in birds caused by HPAI [3]. Moreover, the influenza A virus subtype H5N1, type of HPAI virus (HPAIV), is a threat to the poultry industry and the economy and remains a potential source of pandemic infection in humans [4].
Mx dynamin like GTPase (Mx) proteins are members of the dynamin family of large GTPases. They prevent viral RNA replication by inhibiting the trafficking or activity of viral polymerases [5]. Several previous reports showed that only the asparagine (Asn-AAT) polymorphism at the 631st position triggered antiviral activity, while Mx proteins carrying a serine (Ser-AGT) at that position did not suppress viral growth [6]. In addition, the major histocompatibility complex (MHC) haplotype can affect the antiviral activity of the host [7]. Previous research has shown a significant association of the major histocompatibility complex class I antigen BF2 (BF2) haplotype in chicken MHC class I with resistance or susceptibility to a number of pathogens, including Marek virus [8], and AI virus [9]. Furthermore, chickens with the BF2-B13 haplotype had higher mortality than those that have the BF2-B21 haplotype [9].
One of the key determinants of the severity and outcome of AI virus infection is the regulation of the host innate immune response [10]. Cytokines and chemokines play crucial roles in the balance of the immune system. Previous studies in various animals have shown that tissue damage and host death are caused by cytokine and chemokine dysregulation [11]. In chickens, HPAIV H5N1 induced the excessive expression of cytokines and chemokines in lung tissues [12]. Furthermore, cytokine-cytokine receptor interactions were significantly increased in Fayoumi and Leghorn chicken lines after infection with HPAIV [13]. The regulation of cytokines and chemokines has been shown to be important in host defense against HPAIV.
In this study, we used Ri chickens, a local chicken breed in Vietnam, as an experimental animal [14]. Chickens resistant or susceptible to HPAIV were distinguished by genotyping their Mx and BF2 genes. We infected Ri chickens with HPAIV H5N1 and analyzed gene expression patterns in the lung tissue using high-throughput RNA sequencing. In particular, we analyzed the expression of genes related to cytokine-cytokine receptor interactions between resistant and susceptible Ri chickens.
Fourteen-day-old post-hatching Ri chickens were kept in specific-pathogen-free conditions and observed daily for signs of disease and mortality. The care and experimental use of the chickens were approved by the Ministry of Agriculture and Rural Development of Vietnam (TCVN 8402: 2010/TCVN 8400-26:2014).
For the Mx gene, Ri chickens that have the non-synonymous adenine single nucleotide polymorphism (SNP) at residue 631 were genotyped as resistant while guanine was genotyped as susceptible (Supplementary Figure S1). In addition, for the BF2 gene, those that had the B21 genotype were determined to be resistant and those that had the B13 genotyped were susceptible. Taking these together, HPAIV-resistant Ri chickens had the genotype Mx(A)/BF2 (B21) and HPAIV-susceptible Ri chickens had the genotype Mx(G)/BF2(B13). For HPAIV infection, a total of 10 Ri chickens (five resistant and five susceptible) were inoculated intranasally with the collected allantoic fluid including 104 egg infectious dose (EID50) of A/duck/Vietnam/QB1207/2012 (H5N1) based on the OIE guidelines [15]. All experiments, including chicken management, HPAIV infection, and sample collection, were conducted at the National Institute of Veterinary Research, Vietnam.
At three days post-infection, the lung tissues were collected from five chickens per group, according to the WHO Manual on Animal Influenza Diagnosis and Surveillance. Total RNA from lung tissue was extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
Potentially existing sequencing adapters and low-quality bases in the raw reads were trimmed using Skewer version 0.2.2. The cleaned high-quality reads after trimming the low-quality bases and sequencing adapters were mapped to the reference genome using STAR software version 2.5. Since the sequencing libraries were prepared in a strand-specific manner using the Illumina strand-specific library preparation kit (Illumina Way, San Diego, CA, USA), the strand-specific library option was applied in the mapping process. The gene annotation of the chicken reference genome gg6 from the UCSC genome (https://genome.ucsc.edu) and the corresponding expression values were calculated as fragments per kilobase of transcript per million fragments mapped (FPKM). The differentially expressed genes (DEGs) between the two selected biological conditions were analyzed using the Cuffdiff software in the Cufflinks package version 2.2.1. Assignment to significance was determined using a threshold. The default threshold was set as |fold-change| ≥2 and p-value≤0.05. To gain insight into the biological roles of the genes differentially expressed between the susceptible and resistant lines, a gene set overlapping test was performed between the analyzed DEGs and functionally categorized genes, including the biological processes determined from the gene ontology (GO), Kyoto encyclopedia of genes and genomes (KEGG) pathways, and other functional gene sets using g: Profiler version 0.6.7.
KEGG pathway analysis of the DEGs revealed that most DEGs were related to cytokine-cytokine receptor interactions. Therefore, cytokine-cytokine receptor interactions were analyzed in this study. We used the Search and Color Pathway tool of KEGG Mapper (https://www.genome.jp/kegg/tool/map_pathway2.html) to identify the upregulated or downregulated genes between resistant and susceptible Ri chicken lines. The mutual interactions of the cytokine signaling pathway-related genes were analyzed using the Pathway Studio program (Ariadne Genomics, Rockville, MD, USA).
Quantitative real-time polymerase chain reaction (qRT-PCR) was performed to verify the DEGs obtained from RNA sequencing. Before cDNA synthesis, 2 μg of total RNA (R1D3, R2D3, R3D3, and R5D3 for resistant line and S1D3, S3D3, and S5D3 for susceptible line) was treated with 1.0 U DNase I (Thermo Fisher Scientific, Waltham, MA, USA) to remove potentially contaminating genomic DNA. cDNA synthesis was performed using a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions. The primer sequences of 15 genes and the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were designed using Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) (Table 1). The qRT-PCR was performed in a LightCycler 96 system (Roche, Indianapolis, IN, USA) using the AMPI-GENE qPCR Green Mix Hi-Rox kit (Enzo Life Sciences, Inc., Farmingdale, NY, USA) according to the manufacturer’s recommendations. Relative gene expression was calculated using the 2−ΔΔCt method after normalization with chicken GAPDH [16]. All experiments were performed independently in triplicates.
Statistical analysis was carried out using SPSS software (IBM, SPSS 26.0 for Windows, Chicago, IL, USA), and p<0.05 was considered as a statistically significant expression level using Student’s t-test. All qRT-PCR experiments were replicated independently three times, and the mean±standard error of the mean values for each group were reported.
After H5N1 infection, we observed that chickens had ruffled hair; pulmonary edema, pneumorrhagia, and congestive lung. Among the lung tissue samples obtained from resistant and susceptible Ri chickens three days post H5N1 infection, six samples passed the quality control check and were subject high-throughput RNA sequencing (Table 2).
Table 2 shows the statistics of the raw and clean reads of individual sample transcriptomes after sequence processing and analysis. The six libraries produced approximately 6 GB worth of cDNA sequences per chicken. After data filtering, approximately 21 million clean reads were obtained for each sample from the resistant and susceptible lines.
The number of mapped reads, percentages, and transcripts is shown in Table 3. More than 88.5% of the filtered reads from each library were mapped to the reference genome.
The calculated FPKM values were used to compare the mRNA expression levels between resistant and susceptible Ri chickens (Figure 1). Among the 972 DEGs, 309 genes (32%) were significantly upregulated and 663 genes (68%) were downregulated in susceptible chickens compared to the resistant chicken lines (Supplementary Table S1).
To explore the function of these DEGs, we performed GO and KEGG pathway analyses. Among the 972 DEGs, 114 were associated with biological processes, seven were associated with molecular functions, and three were associated with cellular components (Figure 2; Supplementary Table S2). KEGG pathway analysis revealed six categories of immune-related pathways, including cytokine-cytokine receptor interactions (33 DEGs), nucleotide-binding oligomerization domain like receptor (NOD-like receptor) signaling pathway (27 DEGs), influenza A (26 DEGs), Toll-like receptor signaling pathway (23 DEGs), herpes simplex infection (20 DEGs), and RIG-I-like receptor signaling pathway (11 DEGs) (Figure 3). Most of the DEGs were related to cytokine-cytokine receptor interactions.
Based on the RNA sequencing results and subsequent mapping to the KEGG pathway database, 33 DEGs were involved in cytokine-cytokine receptor interactions (Supplementary Table S3). Among them, 32 genes were significantly downregulated, and one gene was significantly upregulated in susceptible chickens compared to the resistant chicken lines (Figure 4).
For the interaction analysis, 32 DEGs out of 33 genes were identified using the Pathway Studio program (Figure 5). Of these, 28 genes showed strong interactions with each other, but four genes showed no interaction.
To validate the RNA sequencing results, the expression levels of 14 cytokines, chemokines, and receptors of the 33 DEGs between the two chicken lines were quantitatively determined via qRT-PCR (Figure 6). After H5N1 infection, the expression levels of interleukin-1β (IL-1β), IL-6, IL-18, C-C Motif chemokine ligand 4 (CCL4), CCL17, interleukin 8 (CXCL8, IL-8), interleukin 10 receptor subunit beta (IL10RB), atypical chemokine receptor 4 (ACKR4), interferon-γ (IFN-γ), CCL19, and Mx1 were significantly lower in susceptible Ri chickens than in resistant chickens. These results indicate that the expression levels of these genes obtained by qRT-PCR are consistent with RNA-seq results.
In this study, we analyzed the transcriptome profiles of Ri chickens infected with HPAIV H5N1 using RNA sequencing. HPAIV-resistant and HPAIV-susceptible Ri chicken lines were selected based on their Mx and BF2 genotypes and were infected with HPAIV H5N1. RNA sequencing was conducted after infection and 972 DEGs were identified after comparing the transcriptome profiles of lung tissue obtained between resistant and susceptible chickens. KEGG analysis revealed that most of the DEGs were related to cytokine-cytokine receptor interactions.
Avian influenza viral pathogen-associated molecular patterns are recognized by host pattern recognition receptors (PRRs). Viral double-stranded RNA (dsRNA) and cytosine-guanosine oligodeoxynucleotides, which form during the replication of AIV, are recognized by toll-like receptor 3 (TLR3) [17] and TLR21 [18], respectively, through adaptor proteins such as TIR-domain-containing adapter-inducing interferon and myeloid differentiation primary response 88 (MyD88), respectively [19]. These adaptors activate the transcription factor interferon regulatory factor 7 (IRF7) and nuclear factor kappa B (NF-κB) to induce cytokines, chemokines, and IFN-stimulated genes [20]. High expression levels of PRRs, MyD88, IRF7, and NF-κB were previously observed in H5N1-infected chickens [12]. Our results showed that the expression levels of TLR3, TLR21, IRF7, and MyD88 were higher in resistant chickens compared to susceptible chickens (Supplementary Table S1); therefore, we suggest that resistant Ri chickens respond more sensitively to HPAIV H5N1 infection than susceptible Ri chicken lines through increased expression of TLR3, TLR21, IRF7, and MyD88.
Pro-inflammatory cytokines cause inflammation and recruit other immune cells to the site of infection [21]. High expression levels of pro-inflammatory cytokines and chemokines have been found in humans and duck infected with H5N1 [10,11]. Similarly, high expression levels of pro-inflammatory cytokines and chemokines, including IL-1β, IL-6, IL-8, IL-18, CCL4, CCL17, and IFN-γ, were reported in chickens infected with H5N1 [12]. A significant increase in the levels of cytokines and chemokines exacerbates the inflammatory response, leading to apoptosis, multi-organ failure, and host death [22]. However, HPAIV infection was lethal in mice lacking tumor necrosis factor and IL-1 receptors and inhibition of the cytokine response does not protect mice against H5N1 influenza infection [23]. In this study, expression levels of pro-inflammatory cytokines and chemokines such as IL-1β, IL-6, IL-8, IL-18, CCL4, CCL17, IFN-γ, and receptors were upregulated in resistant chickens, compared to susceptible chickens, after HPAIV H5N1 infection (Figure 6). However, the induction of cytokines and chemokines did not cause Ri chickens to die during experimentation. Therefore, we suggest that cytokine and chemokine induction is necessary to protect the host against HPAIV infection.
Type I interferons (IFN-α and IFN-β) trigger the expression of IFN-stimulated genes. IFNs and IFN-stimulated genes can inhibit viral replication by blocking virus entry into the host cells, binding to viral RNA to stop translation, and regulating host antiviral responses [24]. IFN-γ directly or indirectly inhibits viral replication by strictly regulating the production of nitric oxide or interfering with the onset of the RNase L pathway [25]. Moreover, several studies have shown that IFN-stimulated genes have antiviral activity [26, 27]. The Mx gene inhibits the trafficking and activity of viral polymerases [5]. Viperin (RSAD2) inhibits newly synthesized influenza virions [28]. The CCL19, CCL21, and CCR7 chemokine axes are homeostatic [29]. Chicken interferon-inducible 2′-5′-oligoadenylate synthase-like (OASL) and RNase L restrict both viral and cellular RNA, preventing viral genome replication [30]. Wild-type duck OASL inhibits the replication of a variety of RNA viruses in vitro, including the influenza virus [25]. Protein kinase R (EIF2AK2) inhibits the translation of viral mRNAs, including those from influenza A viruses [26]. Interferon-induced proteins of the tetratricopeptide repeats (IFIT) protein family sequester viral nucleic acids [31]. Furthermore, the clinical results were ameliorated in chIFIT5-transgenic chickens after treatment with HPAIV and Newcastle disease virus [27]. As our results showed, although IFN-α and IFN-β were not found in the DEGs, we observed a higher expression of IFN-γ and IFN-stimulated genes (Mx, CCL19, OASL, RSAD2, EIF2AK2, and IFIT5) in the resistant Ri chickens compared to the susceptible ones (Supplementary Table S1). Therefore, we suggest that the antiviral response of resistant chickens is higher than that of susceptible chickens.
In summary, by using RNA sequencing and qRT-PCR, we evaluated the differential expression of genes associated with cytokine-cytokine receptor interactions from the lung tissue of resistant and susceptible H5N1-infected Ri chickens. Interestingly, the expression of PRRs, cytokines, chemokines, and IFN-stimulated genes in resistant chickens were higher than those in susceptible chickens. These results suggest that resistant Ri chickens show a higher antiviral activity compared to susceptible Ri chickens, and this antiviral activity may be related to the expression of antiviral genes. Therefore, we propose that more studies on Ri chickens be done to compare their resistance and susceptibility to HPAIV infections. With this, there will be more studies to support advancing the development of chicken lines with disease resistance genes.
Notes
AUTHOR CONTRIBUTIONS
THV, ADT, KDS, HSL, and YHH conceived and designed the experiments. THV, YJH, ADT, JAL, SYL, KDS, JHC, HVD, HTTT, HSL, and YHH performed the experiments. THV analyzed the data. ADT and YHH contributed reagents, materials, and analytical tools. THV, HYJ, and YHH wrote the manuscript. All authors read and approved the final manuscript.
ACKNOWLEDGMENTS
We thank Department of Biochemistry and Immunology in the National Institute of Veterinary Research, Vietnam for performing animal experiments and collecting samples.
SUPPLEMENTARY MATERIAL
Supplementary file is available from: https://doi.org/10.5713/ab.21.0163
Supplementary Table S1. List of 972 differentially expressed genes selected using the criteria (|fold-change| ≥2) and (p≤0.05) from D3-Resistant and D3-Susceptible chickens
Supplementary Table S2. Gene ontology enrichment of sequence data obtained from the lung tissue of H5N1-resistant and -susceptible chickens, with p<0.05 and Log2-transformation of the normalization of resistant/susceptible ≥2
Supplementary Table S3. List of all genes related to the cytokine-cytokine receptor interaction pathways obtained from the lung tissue of resistant and susceptible H5N1-infected Ri chicken lines (p-value). These data show the changes in gene expression of 190 genes after H5N1 infection
Supplementary Figure S1. Sequencing analysis of the Mx gene in resistant and susceptible Ri chickens
ab-21-0163-suppl.pdf
Figure 1
Heat map of the individual H5N1-infected samples from the lung tissue of resistant and susceptible Ri chickens at three days post-infection. A green color indicates DEGs that have higher expression levels in resistant chickens while a red color indicates DEGs that have lower expression levels in resistant chickens, as calculated from the expression values in log10 (FPKM) units. H5N1, influenza A virus subtype H5N1; DEGs, differentially expressed genes; FPKM, fragments per kilobase of exon model per million reads mapped.
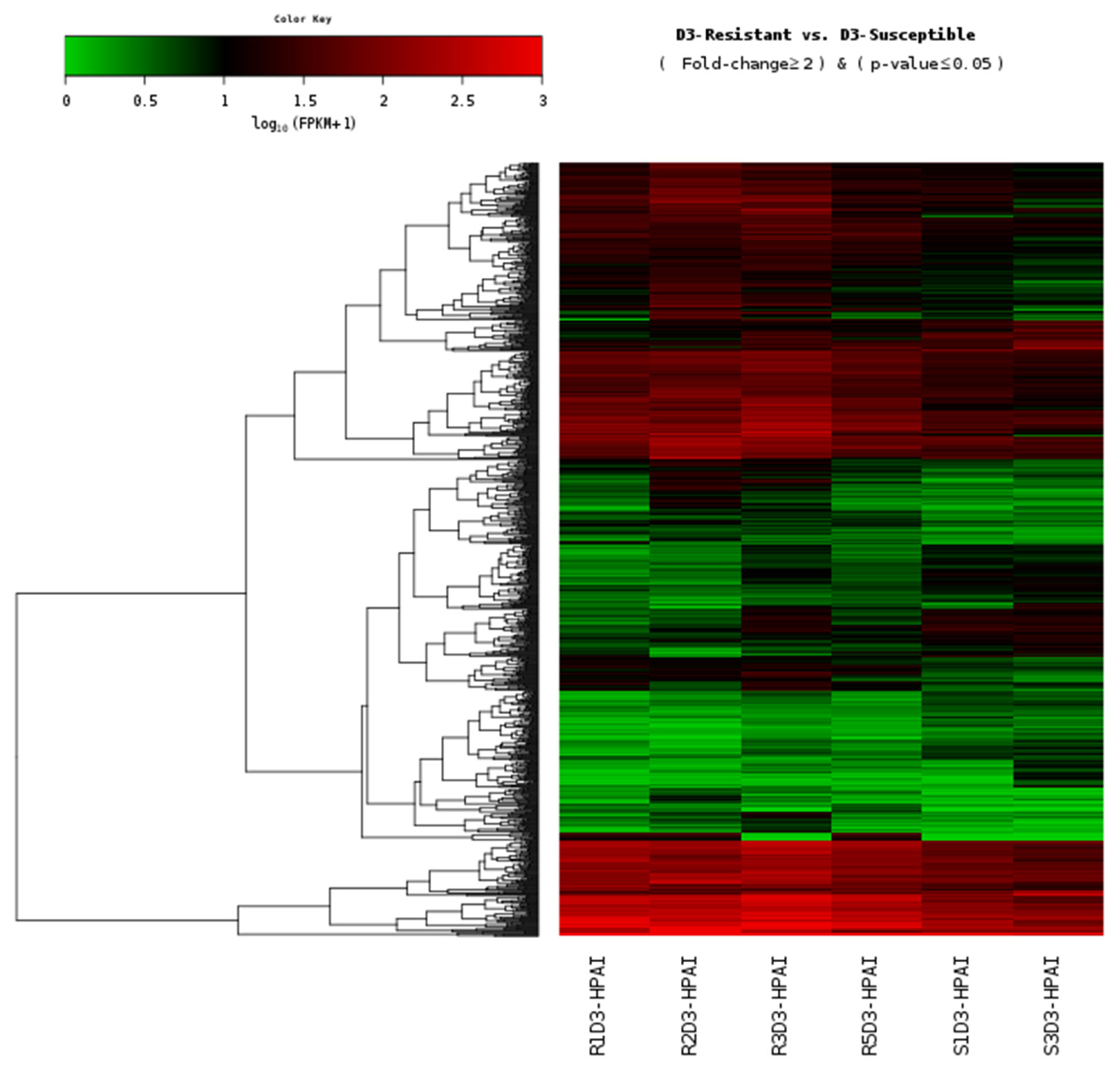
Figure 2
Gene ontology (GO) functional analysis. The enriched biological terms include: (A) Top 10 SEA: GO Biological process, (B) Top 10 SEA: GO Cellular component, and (C) Top 10 SEA: GO Molecular function from the 972 DEG in Ri chicken lines obtained using the criteria (|fold-change| ≥2) ∩ (p≤0.05). GO, gene ontology; SEA, singular enrichment analysis terms; DEG, differentially expressed genes.

Figure 3
Six Kyoto encyclopedia of genes and genomes functional pathways of differentially expressed genes in Ri chickens of the selected differentially expressed genes.
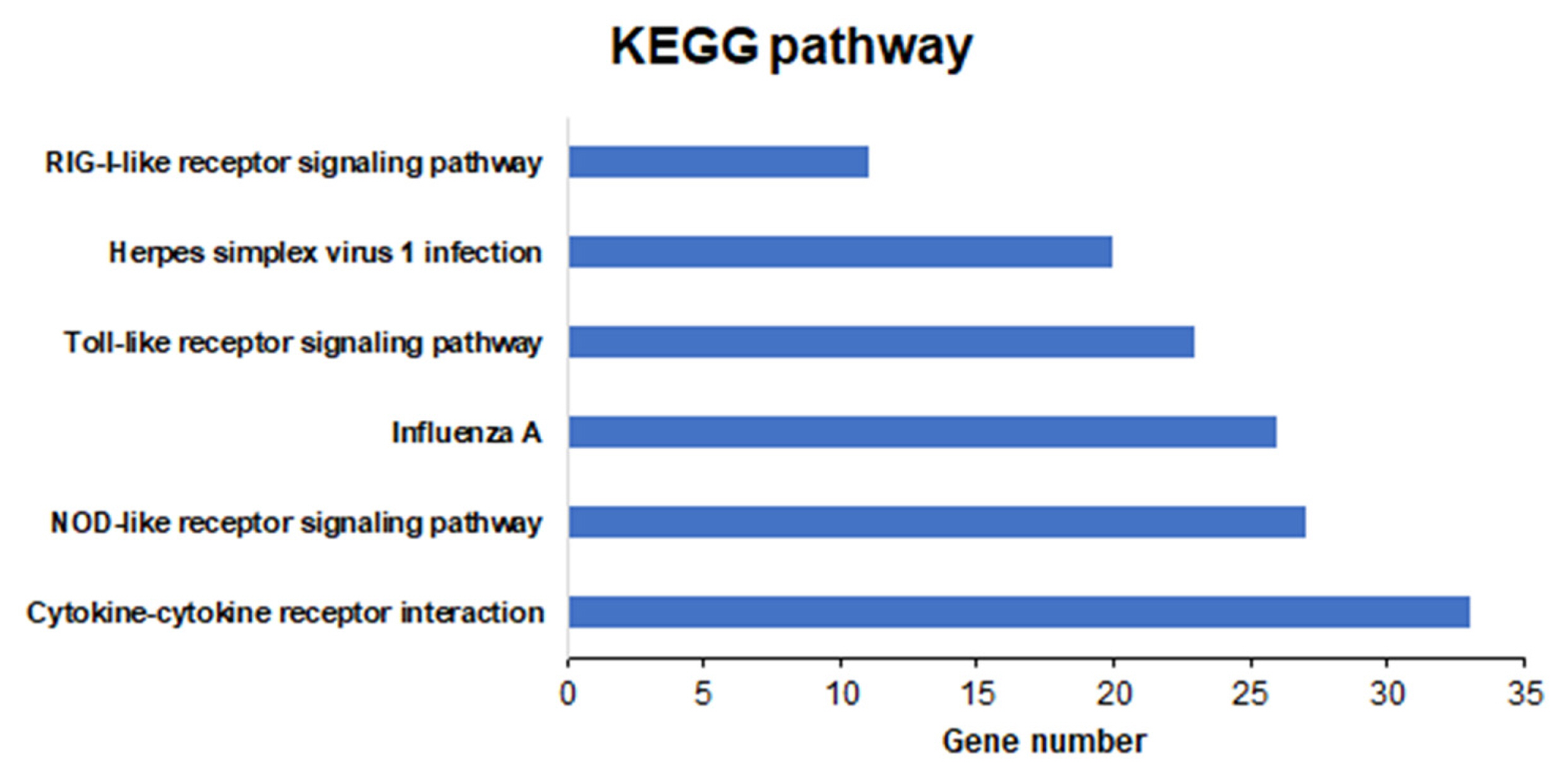
Figure 4
Location of differentially expressed genes associated with cytokine-cytokine receptor interactions in the lungs of resistant H5N1-infected Ri chickens compared with susceptible chickens. The location of 33 significantly different expressed genes were marked. A green color indicates DEGs that have higher expression levels in resistant chickens while a red color indicates DEGs that have lower expression levels in resistant chickens (CXCL14). H5N1, influenza A virus subtype H5N1; DEG, differentially expressed genes.
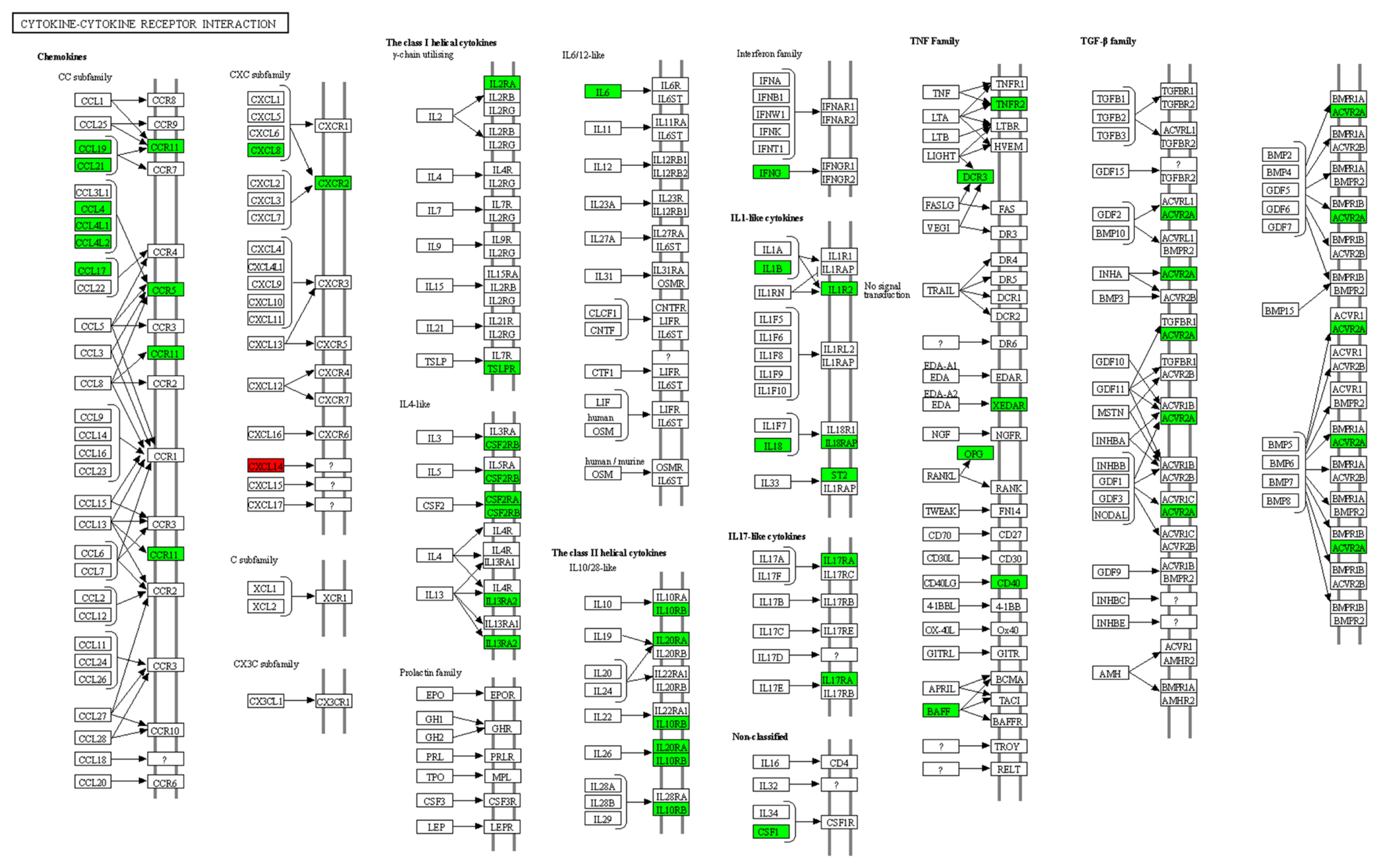
Figure 5
Interactions of the 33 differentially expressed genes in the lungs of Ri chicken lines. This interaction analysis was conducted using the Pathway Studio program.
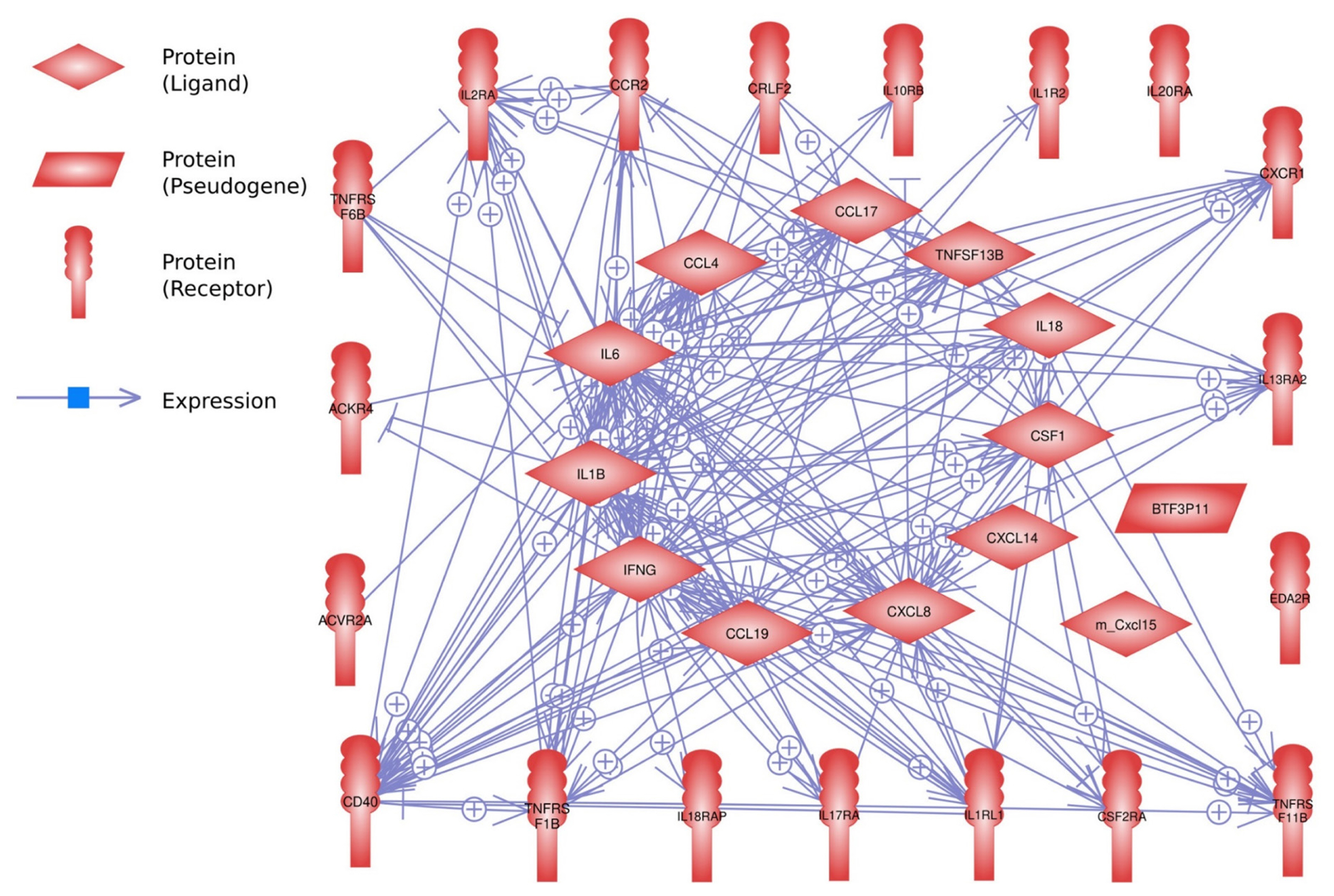
Figure 6
Analysis of the expression of genes associated with cytokine-cytokine receptor interactions in resistant H5N1-infected chickens compared with susceptible H5N1-infected chickens. The expression patterns of these genes were compared between real-time qPCR and RNA sequencing data. Relative quantitation data of qRT-PCR are represented as mean±SEM normalized to GAPDH using the 2−ΔΔCt method. H5N1, influenza A virus subtype H5N1; qRT-PCR, quantitative real-time polymerase chain reaction; SEM, standard error of the mean; GAPDH, glyceraldehyde-3-phosphate dehydrogenase. Data are expressed as mean±SEM of three independent experiments: * p<0.05, ** p<0.01, and *** p<0.001.
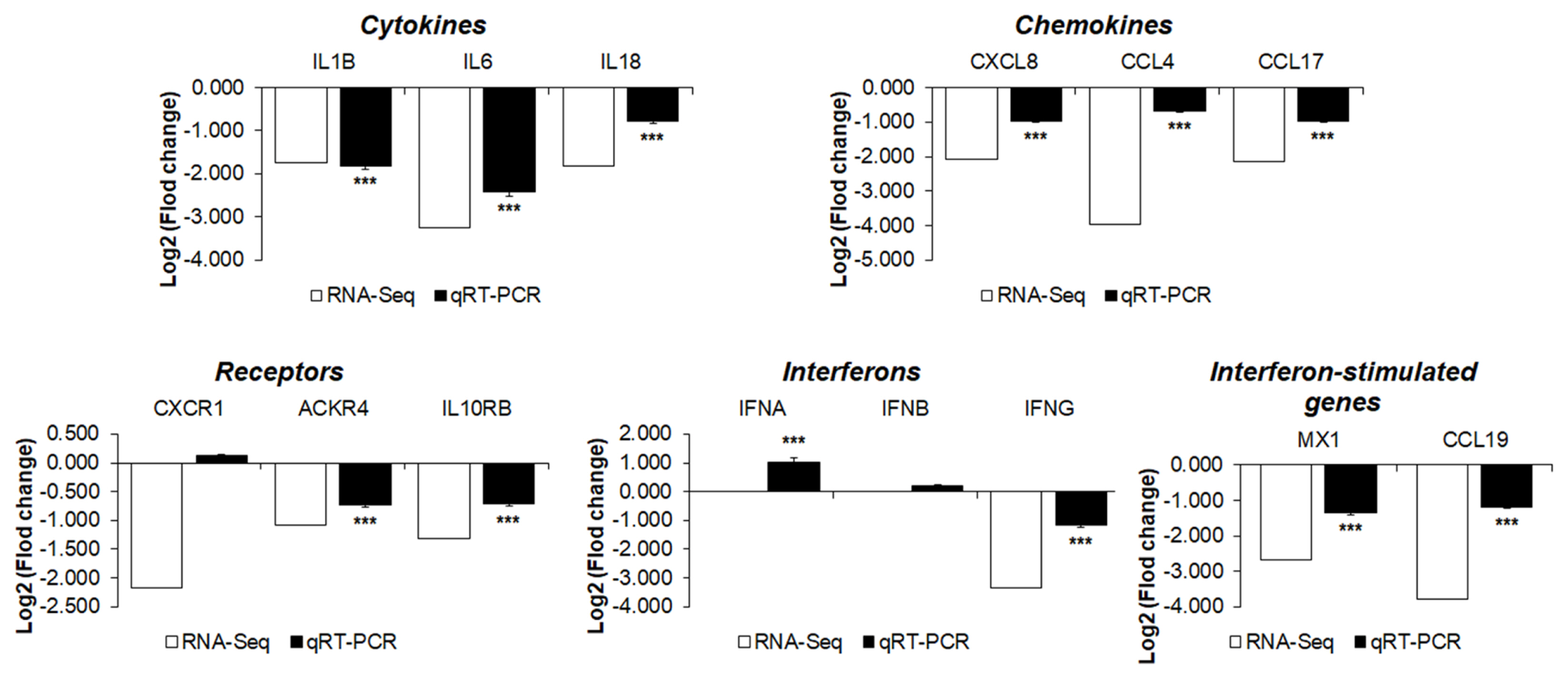
Table 1
Primer sequences used for quantitative real-time polymerase chain reaction
Table 2
Summary of raw reads and clean reads of lung tissue samples obtained from highly pathogenic avian influenza virus-infected chickens from dpi 3 datasets
Table 3
Sequence alignment of the highly pathogenic avian influenza virus-infected samples
REFERENCES
1. Alexander DJ. An overview of the epidemiology of avian influenza. Vaccine 2007; 25:5637–44.
https://doi.org/10.1016/j.vaccine.2006.10.051


2. Chapter O. 10.4 Infection with avian influenza viruses [Internet]. Paris, France: World Organisation for Animal Health; c2001. [2021 July 19]Terrestrial animal health code.
https://www.oie.int/fileadmin/Home/eng/Health_standards/tahc/current/chapitre_avian_influenza_viruses.pdf
3. de Jong JC, Claas ECJ, Osterhaus ADME, Webster RG, Lim WL. A pandemic warning? Nature 1997; 389:554
https://doi.org/10.1038/39218



4. Peiris JM, De Jong MD, Guan Y. Avian influenza virus (H5N1): a threat to human health. Clin Microbiol Rev 2007; 20:243–67.
https://doi.org/10.1128/CMR.00037-06



5. Staeheli P, Pitossi F, Pavlovic J. Mx proteins: GTPases with antiviral activity. Trends Cell Biol 1993; 3:268–72.
https://doi.org/10.1016/0962-8924(93)90055-6


6. Seyama T, Ko J, Ohe M, et al. Population research of genetic polymorphism at amino acid position 631 in chicken Mx protein with differential antiviral activity. Biochem Genet 2006; 44:432–43.
https://doi.org/10.1007/s10528-006-9040-3

7. Jin Y-C, Wei P, Wei X-X, Zhao Z-Y, Li Y. Marek’s disease resistant/susceptible MHC haplotypes in Xiayan chickens identified on the basis of BLB2 PCR-RFLP and BLB2/BF2 sequence analyses. Br Poult Sci 2010; 51:530–9.
https://doi.org/10.1080/00071668.2010.508489


8. Macklin KS, Ewald SJ, Norton RA. Major histocompatibility complex effect on cellulitis among different chicken lines. Avian Pathol 2002; 31:371–6.
https://doi.org/10.1080/03079450220141642


9. Boonyanuwat K, Thummabutra S, Sookmanee N, Vatchavalkhu V, Siripholvat V. Influences of major histocompatibility complex class I haplotypes on avian influenza virus disease traits in Thai indigenous chickens. Anim Sci J 2006; 77:285–9.
https://doi.org/10.1111/j.1740-0929.2006.00350.x

10. De Jong MD, Simmons CP, Thanh TT, et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med 2006; 12:1203–7.
https://doi.org/10.1038/nm1477



11. Saito LB, Diaz-Satizabal L, Evseev D, et al. IFN and cytokine responses in ducks to genetically similar H5N1 influenza A viruses of varying pathogenicity. J Gen Virol 2018; 99:464–74.
https://doi.org/10.1099/jgv.0.001015



12. Ranaware PB, Mishra A, Vijayakumar P, et al. Genome wide host gene expression analysis in chicken lungs infected with avian influenza viruses. PLoS One 2016; 11:e0153671
https://doi.org/10.1371/journal.pone.0153671



13. Wang Y, Lupiani B, Reddy SM, Lamont SJ, Zhou H. RNA-seq analysis revealed novel genes and signaling pathway associated with disease resistance to avian influenza virus infection in chickens. Poult Sci 2014; 93:485–93.
https://doi.org/10.3382/ps.2013-03557


14. Hong Y, Truong AD, Lee J, et al. Exosomal miRNA profiling from H5N1 avian influenza virus-infected chickens. Vet Res 2021; 52:36
https://doi.org/10.1186/s13567-021-00892-3



15. Huprikar J, Rabinowitz S. A simplified plaque assay for influenza viruses in Madin-Darby kidney (MDCK) cells. J Virol Methods 1980; 1:117–20.
https://doi.org/10.1016/0166-0934(80)90020-8


16. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001; 25:402–8.
https://doi.org/10.1006/meth.2001.1262


17. Yong Y-H, Liu S-F, Hua G-H, et al. Goose toll-like receptor 3 (TLR3) mediated IFN-γ and IL-6 in anti-H5N1 avian influenza virus response. Vet Immunol Immunopathol 2018; 197:31–8.
https://doi.org/10.1016/j.vetimm.2018.01.010


18. Abdul-Cader MS, Ahmed-Hassan H, Amarasinghe A, et al. Toll-like receptor (TLR) 21 signalling-mediated antiviral response against avian influenza virus infection correlates with macrophage recruitment and nitric oxide production. J Gen Virol 2017; 98:1209–23.
https://doi.org/10.1099/jgv.0.000787


19. Wei L, Jiao P, Yuan R, et al. Goose toll-like receptor 7 (TLR7), myeloid differentiation factor 88 (MyD88) and antiviral molecules involved in anti-H5N1 highly pathogenic avian influenza virus response. Vet Immunol Immunopathol 2013; 153:99–106.
https://doi.org/10.1016/j.vetimm.2013.02.012


20. Barjesteh N, Abdelaziz KT, Sharif S. The role of IRF7 and NF-κB pathways in the induction of antiviral responses in chicken tracheal epithelial cells following exposure to TLR3 and 4 ligands. J Immunol 2016; 196:Suppl 1216–8.


21. Nturibi E, Bhagwat AR, Coburn S, Myerburg MM, Lakdawala SS. Intracellular colocalization of influenza viral RNA and Rab11A is dependent upon microtubule filaments. J Virol 2017; 91:e01179–17.
https://doi.org/10.1128/JVI.01179-17



22. Cheung C, Poon L, Lau A, et al. Induction of proinflammatory cytokines in human macrophages by influenza A (H5N1) viruses: a mechanism for the unusual severity of human disease? The Lancet 2002; 360:1831–7.
https://doi.org/10.1016/S0140-6736(02)11772-7

23. Salomon R, Hoffmann E, Webster RG. Inhibition of the cytokine response does not protect against lethal H5N1 influenza infection. Proc Natl Acad Sci USA 2007; 104:12479–81.
https://doi.org/10.1073/pnas.0705289104



24. De Silva Senapathi U, Abdul-Cader MS, Amarasinghe A, et al. The in ovo delivery of CpG oligonucleotides protects against infectious bronchitis with the recruitment of immune cells into the respiratory tract of chickens. Viruses 2018; 10:635
https://doi.org/10.3390/v10110635



25. Rong E, Wang X, Chen H, et al. Molecular mechanisms for the adaptive switching between the OAS/RNase L and OASL/RIG-I pathways in birds and mammals. Front Immunol 2018; 9:1398
https://doi.org/10.3389/fimmu.2018.01398



26. Garcia MA, Gil J, Ventoso I, et al. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol Mol Biol Rev 2006; 70:1032–60.
https://doi.org/10.1128/MMBR.00027-06



27. Rohaim MA, Santhakumar D, Naggar RFE, et al. Chickens expressing IFIT5 ameliorate clinical outcome and pathology of highly pathogenic avian influenza and velogenic newcastle disease viruses. Front Immunol 2018; 9:2025
https://doi.org/10.3389/fimmu.2018.02025



28. Wang X, Hinson ER, Cresswell P. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe 2007; 2:96–105.
https://doi.org/10.1016/j.chom.2007.06.009


29. Förster R, Davalos-Misslitz AC, Rot A. CCR7 and its ligands: balancing immunity and tolerance. Nat Rev Immunol 2008; 8:362–71.
https://doi.org/10.1038/nri2297


30. Tag-EL-Din-Hassan HT, Morimatsu M, Agui T. Functional analysis of duck, goose, and ostrich 2′-5′-oligoadenylate synthetase. Infect Genet Evol 2018; 62:220–32.
https://doi.org/10.1016/j.meegid.2018.04.036


31. Pichlmair A, Lassnig C, Eberle C-A, et al. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat Immunol 2011; 12:624–30.
https://doi.org/10.1038/ni.2048








 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Supplement
Supplement Print
Print





