INTRODUCTION
There is a common practice in recent years to anaerobically ferment high-moisture and perishable agricultural and food by-products with dry feeds as total mixed ration (TMR) silages [
1–
3]. The efficient utilization of by-products helps to develop new feed resources for ruminants, and the ensiling process results in highly enhanced preservation [
4], facilitating the transportation and flexible utilization of TMR without the occurrence of aerobic deterioration for a relatively long period [
5].
Ensiling is a complex process that involves the interaction of plant enzymes and numerous microbial species, ultimately changing the biochemistry of silage [
6]. Hydrolysis of protein is expected to occur during the whole fermentative stage, and it has been well accepted a result of the plant proteinases and the microbial activities in silages. Plant proteins can be degraded into oligopeptides, free amino acids, ammonia and other forms of non-protein nitrogen in silages (NPN) [
7]. The utilization of silage NPN in rumen is often less efficient, and the inefficient NPN metabolism can lead to the nitrogen loss to the environment, which is now an increasing concern on silage-based diets in livestock production [
8].
Proteolysis in ensiled forage has been mainly considered a result of the plant proteolytic enzymes [
7,
9]. So far, the plant proteinases and peptidases have been principally clarified, and their relative contributions to the formation of various NPN compounds have been well demonstrated in silages. According to Guo et al [
10] and Tao et al [
11], protein hydrolysis mainly resulted from the plant exo- and endopeptidases, and the principle exo- and endopeptidases hydrolyzing the forage protein were di-, tripeptidyl- and carboxypeptidases, and metallo and cysteine peptidases, respectively. McKersie and Buchanan-Smith [
12] also demonstrated that carboxypeptidase, aminopeptidase, and acid proteinase were important in the degradative process of protein in ensiled alfalfa. In addition to the well-accepted effects of the plant proteinases, microbial activities were also reported to participate in the proteolysis during ensiling. According to Winters et al [
13], microbial proteinases played an important role in the metabolism of amino acids in the ensiled ryegrass, and Heron et al [
14] also pointed out that the ammonia and amines were the largely end products of microbial proteinases. Tao et al [
15] investigated the effects of epiphytic bacteria and exogenous lactic acid bacteria (LAB) on the proteolysis in alfalfa silage, and concluded that both the aerobic bacteria and LAB species could affect the formation of NPN constitutes during the fermentative period. However, particular emphasis has been made in studies on the reactions involved in protein hydrolysis in regular silages, whereas the degradation of protein and its metabolism did not receive the same interest in TMR silages. In this experiment, we described the proteolytic enzymatic characterization and the dominant proteolytic bacteria succession in TMR silages during ensiling. Different classes of proteinases in the proteolytic isolates were also investigated in our study to elucidate the possible role of these specific strains in the hydrolysis of protein during anaerobic fermentation in TMR silages.
DISCUSSION
One of the most significant processes occurring during ensiling is the enzymatic degradation of protein to the NPN forms in silages. Even in well-preserved silages, approximately 50% degradation of protein may take place [
24,
25]. Alfalfa is particularly susceptible to proteolysis, because the proteinase activity is higher in this crop than in other grasses or legumes [
26]. So far, studies have been mainly focused on the proteolysis in silages, especially in alfalfa silage, while the degradation of protein and its mechanism has not been well demonstrated in TMR silages. This experiment showed the intensities of proteolysis, activities and characterizations of proteinases, and the dominant proteolytic microorganisms in A-TMR and L-TMR silages. Furthermore, it was the first attempt to determine the contributions of the predominating proteolytic species to the hydrolysis of protein in TMR silages, and might aid to elucidate the mechanism of proteolysis from a microbial perspective.
Both plant enzyme and microbial activity are factors responsible for the extensive breakdown of protein in silages. It has been well accepted that the initial hydrolysis is mediated by plant enzymes, and the subsequent transformation of amino acids to ammonia and amines is mostly brought about by the activities of proteolytic microorganisms [
7]. However, the above statement that proteolytic activity during ensiling is largely attributed to plant enzymes seems not to be entirely suitable for TMR silages in this experiment. Soybean curd residue and other food by-products, such as brewers’ grains [
1], green tea ground, and apple pomace [
2] which are commonly utilized to replace the commercial feedstuffs in TMRs might have lost their enzymatic activities through the procedures of food processing and manufacturing, and the enzymes in dry feeds or hays might also be inactive because of their low levels of the moisture contents. Thus, in this experiment, it could be assumed that the breakdown of protein in TMR silages was predominantly a microbial process.
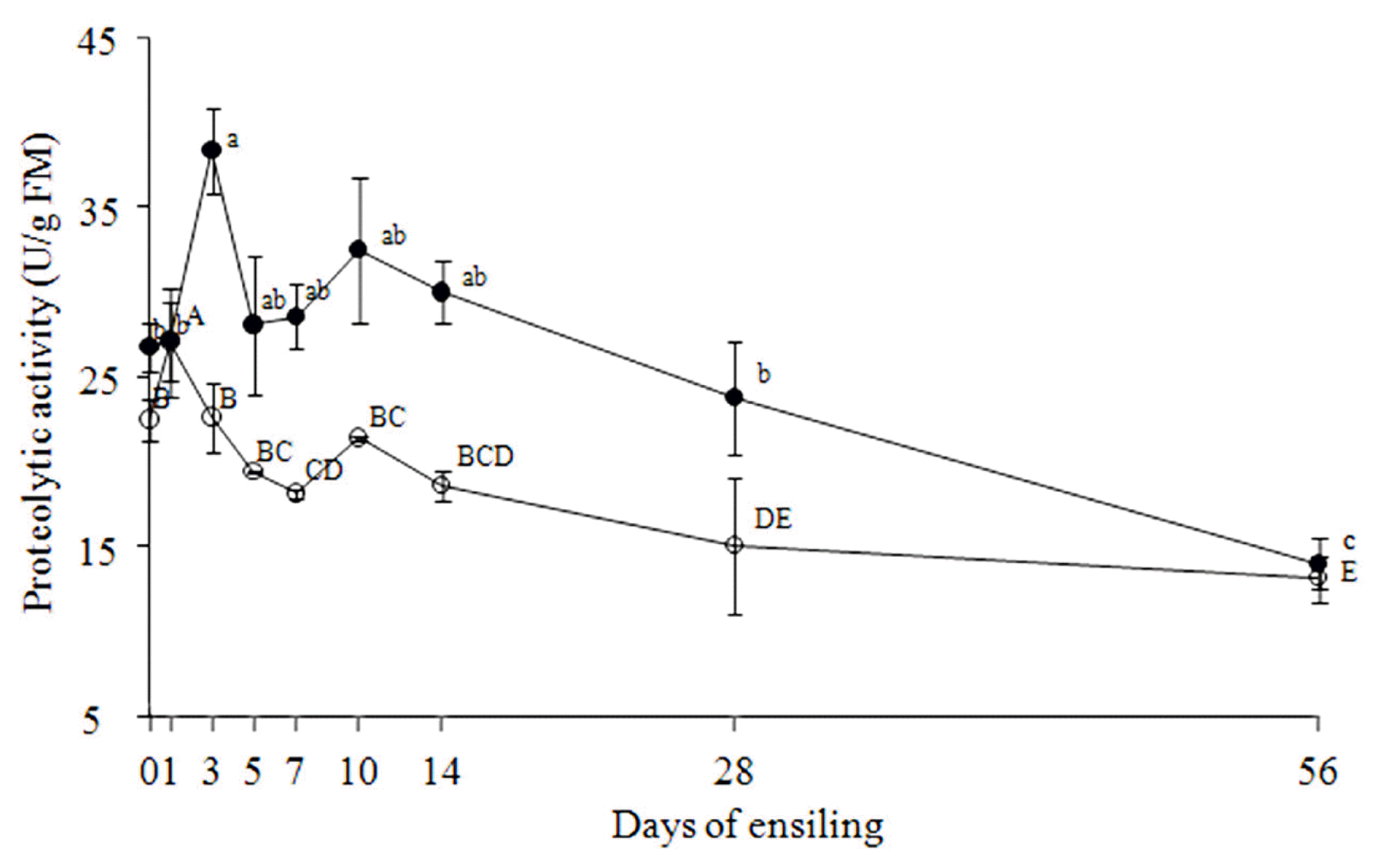
Total activities of the microbial proteinases in both TMR silages exhibited dynamic changes during the ensiling process. Proteinases produced by microorganisms are complex and their activities can be affected by many factors, such as microorganism strain, substrate, temperature, and pH value. In this experiment, the increases of the proteinase activities during initial ensiling stage could be largely due to the temporary proliferation of the proteolytic species of aerobic bacteria, and the following decreases might be related to the inhibition of aerobic bacteria and the multiplication of LAB under the anaerobic and slightly acidic environments. Tao et al [
15] once reported the effects of epiphytic microorganisms and exogenous LAB on the extent of proteolysis in alfalfa silages. Control silage had significantly higher concentrations of peptide-N, FAA-N and NH
3-N than the silage inoculated with exogenous LAB after UV radiation, and this result roughly indicated that the epiphytic aerobic bacteria largely contributed to the formation of NPN compositions during ensiling period. Except for the increased population of the proteolytic LAB strains and their relatively low proteinase activities, the predominantly reduced pH value might also be an important factor leading to the inhibited proteolytic activities in TMR silages when ensiling prolonged. McKersie and Buchanan-Smith [
27] reported that the activity of the native proteinase against azocasein (pH 5.5) decreased with the reduced pH value in alfalfa silage, and the proteinase activity retained only 30% of its original activity at 21 d after the anaerobic fermentation.
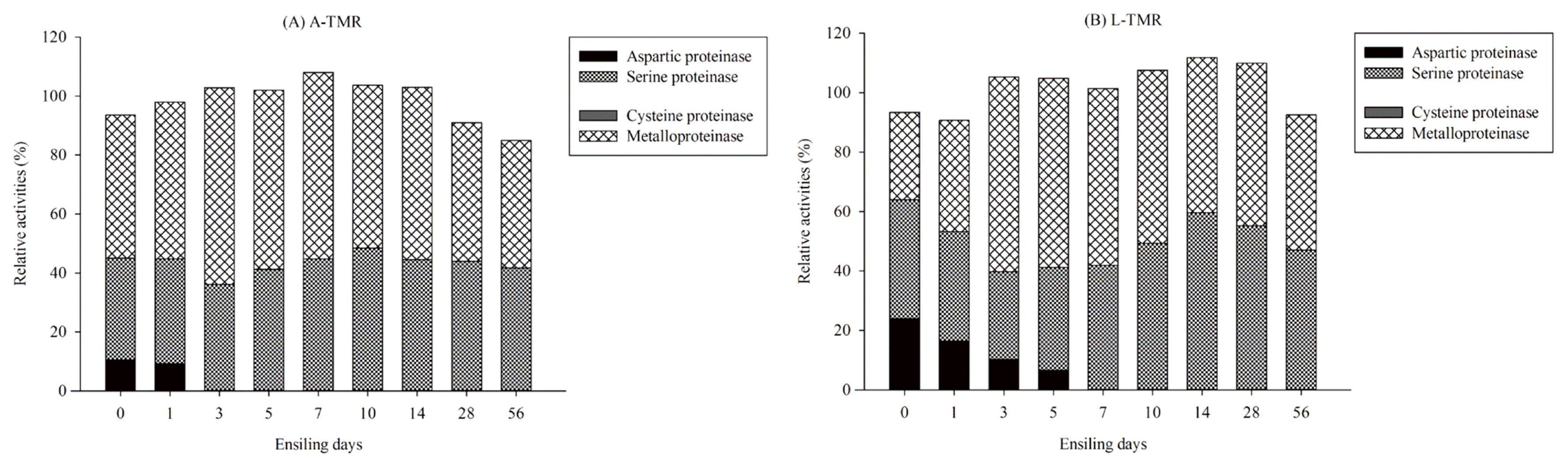
Proteinase can be classified according to the active amino acid residues at the enzyme reaction point, namely, aspartic, serine, cysteine and metallo proteinases, and different classes of proteinases make different contributions to the hydrolysis of protein. According to Guo et al [
10], aspartic and cysteine proteinases mainly contribute to the degradation of protein into oligopeptides, serine and metallo proteinases principally hydrolyze peptides into free amino acids, and the formation of NH
3-N is largely due to metalloproteinase in alfalfa silages. In this experiment, the above four classes of proteinases were determined with the application of their specific inhibitors. Results showed that the serine and metallo proteinases accounted for the majority of the microbial proteinases in both TMR silages, and L-TMR silage exhibited significantly higher values of the absolute activities of serine and metallo proteinases than A-TMR silage throughout the whole ensiling period. This was consistent with the result that L-TMR silage obtained higher intensities of the degradation of peptides and higher concentrations of free amino acids and NH
3-N during ensiling in our study. The microbial aspartic and cysteine proteinases were either significantly suppressed during the initial fermentation stage or totally inactive throughout the whole ensiling period, and this might be a reason that TMR silages exhibited significantly lower degrees of protein breakdown than regular silages, and the gradually decreased activities of the aspartic proteinases in both TMR silages might lead to the phenomenon that the rising rates of the peptide concentrations progressively declined with the increased fermentative duration. In addition, the initial hydrolysis of protein and the subsequent degradation of peptides into free amino acids might also result from the exopeptidases activities in TMR silages. According to Tao et al [
11], the endo- and exopeptidases both contribute to the protein hydrolysis and the tripeptidyl-peptidase was a principle exopeptidase for protein degradation in alfalfa silage. So, there is also a necessity for another determination of the microbial exopeptidases in TMR silages in a further study to conclusively link the observed proteolysis with the specific bacteria species which have the capability of producing exopeptidases during the ensiling process.
Casein was used as the single nitrogen source in the selective media for the isolation of proteolytic strains in our study. However, it has been confirmed that these selective media are not particularly efficient for selecting proteolytic microorganisms [
28], and a number of proteolytic isolates might fail to clear the casein-containing medium, presumably because their cell-bound proteinase cannot diffuse through the medium. So in this experiment, strains without clear haloes were also isolated and purified for their further analysis of the proteinase-producing abilities. In the unensiled A-TMR, proteolytic strains of
C. flaccumfaciens and
C. Citreum were isolated from 10
−3 dilutions of the leaching solutions, and these strains were supposed to be the most dominant proteolytic isolates during the initial fermentation stage. The proteinases in
C. flaccumfaciens and
C. Citreum were mainly of the metalloproteinase type and this was in accord with the result that metalloproteinase occupied most of the total microbial proteinases in the initial A-TMR silage. In the unensiled L-TMR,
P. borealis,
P. turicensis, and
P. xylanexedens were isolated as the most dominant proteolytic strains in this experiment. Their proteinases mainly had metalloproteinase characteristics, and aspartic proteinase was also weakly active in their proteolytic systems. In our study, the total microbial proteinase reached its maximal activity at 3 d, with metalloproteinase taking the majority of the proteinases in L-TMR silage, and these results were consistent with the dynamics of the populations of the proteolytic
Paenibacillus strains during the initial fermentation. The relative activity of the aspartic proteinase gradually decreased even with the proliferation of
Paenibacillus strains in L-TMR silage, this might be a result from the significantly reduced pH values, since the aspartic proteinase is a neutral protease, and might be totally inactive at pH 5 [
9]. There are very limited reports about the proteolytic activity expressed by members of the genus
Paenibacillus. Hang et al [
29] isolated a proteolytic strain of
Paenibacillus spp. from raw yak milk, and discovered that the enzyme activity was completely inhibited by ethylenediaminetetraacetic acid, indicating it was a metalloproteinase. The extracellular proteinase profiles of the other two
Paenibacillus species,
Paenibacillus peoriae and
Paenibacillus polymyxa were also analyzed. The results showed that both the proteinases were at neutral-alkaline pH range, and of metalloproteinase class [
30].
B. amyloliquefaciens and
B. methylotrophicus were the most active isolates in both TMR silages of this experiment. Their activities were mainly with serine and metallo proteinases characteristics, and similar results were also obtained by Sai-Ut et al [
31] before. Proteolytic strains of other
Bacillus species were isolated in lower orders of magnitudes. Since
Bacillus species are well known for their capability of producing the extracellular alkaline proteinases, with their activities remarkably decreased at the acidic pH range, the contributions of the genus
Bacillus to proteoylsis could be definitely weakened with the increased fermentative duration.
Since LAB have a complex proteolytic system capable of converting protein to free amino acids [
32], the proteolytic activity during the later ensiling phase could be due to the proteolytic strains of the genus
Lactobacillus in TMR silages. Highly proteolytic strains of
Lactobacillus helveticus,
Lactobacillus paracasei (
L. paracasei) subsp.
paracasei,
Lactobacillus acidophilus,
Lactobacillus casei (L. casei),
Lactobacillus buchneri (
L. buchneri), and
Lactobacillus delbrueki subsp.
bulgaricus have been identified previously [
33]. So far, many studies have characterized the amino acids changes mediated by LAB in the mature silages, such as the metabolisation of arginine by strains of
E. faecium,
Enterococcus faecalis,
L. buchneri,
L. plantarum, and
L. casei [
34], and the fermentation of serine and proline, respectively, by strains of
L. plantarum and
L. paracasei [
13]. There are few studies examining the effects of LAB proteinases to the initial hydrolysis of protein to peptides in TMR silages.
L. plantarum,
P. acidilactici, and
E. faecium were isolated as the proteolytic LAB strains in our study. Proteinase activities in proteolytic strains of
L. plantarum are relatively low, and the inhibitor study suggested that the
L. plantarum proteolytic enzyme was a serine proteinase. The proteolytic activities in
L. plantarum have been reported helping in casein degradation in milk [
35], and the characterization of serine proteinase were also reported in a
L. plantarum proteinase by Hegazi and Abo-Elnaga [
36]. The proteinase produced by a proteolytic strain of
P. acidilactici has been studied previously. The results showed that the proteinase was an acidic proteinase [
37], and this confirmed the result that
P. acidilactici contributed to the proteolysis in both mature TMR silages with pH values of approximately 4.2 in this experiment. Proteoyltic activities of genus
Enterococci were mainly described in milk products. The maximal degradation of milk protein was achieved at pH 6.5 and at 42°C for
E. faecium, and the proteinase largely exhibited metalloproteinase characteristics according to El-Ghaisha et al [
38]. In our study, the proteinases in the determined
E. faecium strains mostly belonged to the metalloproteinase class, and serine proteinase was also active in their proteolytic systems. These results were mainly inconsistent with previous studies, and the difference between the results may be due to the different proteolytic strains of
E. faecium in our study relative to the previous reported researches. Our study indicated that, with increased fermentative duration, the proteolytic aerobic bacteria strains were substituted by the proteolytic LAB, and the serine and metallo proteinases in these proteolytic strains played leading roles in the hydrolysis of protein in TMR silages. This work throws new light on the mechanisms of protein hydrolysis occurring in TMR silage from a microbial perspective, and further studies are needed to isolate the strains with exopeptidase activities and to evaluate the effects of these proteolytic species on the formation of NPN in TMR silages. There is also necessity to establish the significance of the endo- and exo- peptidases inhibitors on the protein quality in TMR silages, in the hope that it can provide some useful information for commercial development of proteolysis-inhibiting additives for TMR silages.








 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print





