 |
 |
| Anim Biosci > Volume 30(4); 2017 > Article |
|
Abstract
Objective
Manipulating the fermentation to improve the performance of the ruminant has attracted the attention of both farmers and animal scientists. Propionate salt supplementation in the diet could disturb the concentration of propionate and total volatile fatty acids in the rumen. This study was conducted to evaluate the effect of calcium propionate supplementation on the ruminal bacterial community composition in finishing bulls.
Methods
Eight finishing bulls were randomly assigned to control group (CONT) and calcium propionate supplementation (PROP) feeding group, with four head per group. The control group was fed normal the total mixed ration (TMR) finishing diet, and PROP group was fed TMR supplemented with 200 g/d calcium propionate. At the end of the 51-day feeding trial, all bulls were slaughtered and rumen fluid was collected from each of the animals.
Results
Propionate supplementation had no influence the rumen fermentation parameters (p>0.05). Ruminal bacterial community composition was analyzed by sequencing of hypervariable V3 regions of the 16S rRNA gene. The most abundant phyla were the Firmicutes (60.68%) and Bacteroidetes (23.67%), followed by Tenericutes (4.95%) and TM7 (3.39%). The predominant genera included Succiniclasticum (9.43%), Butyrivibrio (3.74%), Ruminococcus (3.46%) and Prevotella (2.86%). Bacterial community composition in the two groups were highly similar, except the abundance of Tenericutes declined along with the calcium propionate supplementation (p = 0.0078).
Ruminants harbor a diverse microbial community to break down dietary plant material to fermentation, and the host requires ruminal fermentation end products for body maintenance and growth, and milk production. Volatile fatty acids (VFAs), absorbed by the host across the rumen wall, serve as the major carbon and energy sources for ruminants. Acetic, propionic, and butyric acids normally make up 95% of total VFA present in the rumen, of which propionic acid accounts for between 25% and 44% [1]. As known, beef cattle can be fed with a high concentrate diet, mainly composed of maize or barley, to improve fat deposition in the finishing period. The feeding strategy could aim to increase propionate concentration, and reduce the acetate/propionate (A/P) ratio in the rumen. Propionate produced in rumen is the main precursor required for glucose synthesis in the liver [2], which supplies 32% to 73% of glucose demands [3].
Manipulating the fermentation to improve the performance of the ruminant has attracted the attention of both farmers and animal scientists. To date, most reports of propionate salt supplementation in ruminant diets focused on performance, such as dry matter digestibility [4ŌĆō7], carcass index [8,9], and milk and protein yield [7,10,11]. In ruminants, calcium propionate supplementation increased the concentration of propionate in the rumen [5,6,8]. Providing propionate to ruminants may influence microbial dynamics due to the changing of the propionate concentrate in the rumen. However, few studies have reported the effect of propionate supplementation on ruminal microbial diversity in finishing stage. So, it is necessary to understand the bacterial community composition changes in response to calcium propionate supplementation. In the present study, we hypothesize that calcium propionate addition may disturb the ruminal microbial ecology in finishing bulls. Therefore, our objective was to investigate the effect of propionate supplementation on ruminal bacterial community composition in finishing bulls by using pyrosequencing of 16s RNA gene.
Animals used in this study were handled in strict accordance with the Regulations for Laboratory Animals of Beijing. The protocol was approved by the Animal Welfare Committee of the China Agricultural University (Permit Number: DK1103). Healthy Japanese black Wagyu bulls (n = 8), with an average body weight of 630┬▒15 kg, were randomly divided into two groups: control feeding group (CONT), which received the normal diet, and calcium propionate supplementation feeding group (PROP), which received the normal diet with calcium propionate supplement. The basal diet consisted of 52.35% corn grain, 25.00% silage corn stalks, 16.00% brewerŌĆÖs grains, 4.00% cotton seed meal, 1.00% sodium bicarbonate, 0.60% limestone, 0.50% trace mineral-salt, 0.25% magnesium oxide, 0.10% calcium phosphate and 0.20% vitamin premix on a dry matter basis. The concentrate component of calcium propionate was changed at the beginning on d 0, and in a stepwise fashion to reach the desired amount of calcium propionate supplementation (200 g/d). Animals were fed twice daily at approximately 0800 and 1700. Calcium propionate (99.26% purity, Dongxin company, Thenghua, China) was mixed with the total mixed ration (TMR) in the PROP group. Finishing bulls were individually housed in tie-stall barns, and allowed ad libitum access to water.
At the conclusion of the 51 day feeding period, all bulls were slaughtered to collect rumen contents. Animals were loaded and transported to a commercial slaughter house. Efforts were made to minimize the suffering of animals. The finishing bulls were restrained in an upright position with their head held fast and the neck exposed in a suitable position for the incision of the throat. The knife used for bulls has a long, extremely sharp, and undamaged blade. The intention was to produce the immediate outpouring of blood by severing both jugular veins and both carotid arteries.
After bulls were slaughtered, ruminal digesta from the dorsal, central, and ventral regions of the rumen were collected to form one composite sample, and then the contents from each bull were strained via four layers of cheesecloth, and the liquid fraction was placed into a sealed 50-mL conical tube. Sampling was accomplished as quickly as possible. Rumen samples were then frozen in liquid nitrogen prior to storage at ŌłÆ80┬░C until genomic DNA was extracted. For the ruminal fermentation parameters, pH value of the rumen contents was measured immediately with a portable pH meter coupled to a glass electrode (Model PHS-3C, Shanghai Leici Scientific Instrument Co., Ltd., Shanghai, China). Rumen samples were centrifuged at 8,000├Śg, 4┬░C for 15 min, collecting the supernatant fluid. Ammonia nitrogen (NH3-N) was analyzed by colorimetric procedure [12], VFA concentrations were determined with a gas chromatograph Agilent 6890 (Agilent Technologies Inc., Santa Clara, CA, USA) equipped with an HP-INNO wax capillary column (30 m├Ś0.32 mm) [13].
Total genomic DNA from 200 ╬╝L of frozen rumen samples was extracted using TIANGEN TIANamp Stool DNA Kit (Tiangen Biotech Co., Ltd., Beijing, China) as previously described [14]. Briefly, cells were disrupted by combined beat-beating and manufacturers recommended methods. DNA was precipitated with 2 volumes of 30% (wt:vol) polyethylene glycol, washed with 70% (vol:vol) ice-cold ethanol and eluted in 100 ╬╝L elution buffer (10 mM Tris, pH 8.5 with HCl). RNA was digested with 50 ╬╝g RNase A, and DNA was subsequently cleaned up and eluted in 50 ╬╝L Tris-HCl+ethylenediaminetetraacetic acid. The yield and purity of the extracted DNA were assessed with a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific Inc., Wilmington, DE, USA).
Pyrosequencing targeted the hypervariable V3 region of the 16S rRNA gene was amplified from genomic DNA with universal eubacterial primers F357 (5ŌĆ▓-CCTACGGGAGGCAGCAG-3) and R534 (5ŌĆ▓-ATTACCGCGGCTGCTGG-3ŌĆ▓). The amplification conditions consisted of an initial denaturation at 98┬░C or 3 min; 30 cycles of denaturation at 98┬░C for 10 s, annealing at 58┬░C for 30 s, elongation at 72┬░C for 30 s, and a final 5 min extension at 72┬░C. Amplicons were selected on 2% agarose gels on E-Gel Size Select Agarose Gel and then purified with Agenc AMPure XP Reagent. The purified DNA was quantified with Quant-iTTM Technology (Life Technologies, Inc., Carlsbad, CA, USA) applying Quant-iTTM dsDNA Broad-Range Assay Kit. Agilent 2100 BioanalyzerTM with the Agilent High Sensitivity DNA Kit (Agilent Technologies, Inc., Santa Clara, CA, USA) was used to analyze library sizes and molar concentrations. The emulsion polymerase chain reaction (PCR) was performed using the Ion OneTouchTM 200 Template Kit v2 DL (Life Technologies, Inc., USA) according to the manufacturerŌĆÖs instructions. Sequencing of the amplicon libraries was conducted on a 318 chip with the Ion Torrent Personal Genome Machine (PGM) system using the Ion PGMTM. Sequencing 300 kit (Life Technologies, Inc., USA).
The Ion Torrent sequencing data were analyzed using the QIIME pipeline v1.7 [15]. Raw reads were first decoded based on 8 bp sample-specific barcodes. Depending upon appropriate fragment size for V3 PCR (150 to 200 bp), bases after position 200 were trimmed and reads shorter than 150 bp were removed. Then reads with a quality score <25 were removed with NGS QC Toolkit and only sequences without ambiguous characters were included in the analysis. To calculate downstream diversity determination (alpha and beta diversity), all samples were subsampled to equal size 12,000 before comparing the subsequent bacterial communities. The sequences were clustered into operational taxonomic units (OTUs) at the 97% sequence identity level and the most abundant sequence of each OTU was chosen as a representative. The sequence of each OTU was assigned to the lowest possible taxonomic rank with QIIME; a reference dataset from the Greengenes database (gg_13_5_otus) was used. Alpha diversity indices (observed species, Chao, and Shannon) were generated with the QIIME pipeline, and ╬▓ diversity (i.e., diversity between groups of samples) was used to create principal coordinate analysis (PCoA) plots using unweighted and weighted UniFrac distances. The two-dimensional hierarchical clustering heatmap was drawn based on the number of reads of each OTU using pheatmap package in R software (version 3.1.0), OTUs containing reads <200 were filtered.
Relative quantification method was used as reported previously [16,17]. The abundance of twelve specific rumen bacteria were quantified using quantitative real-time polymerase chain reaction (qPCR) assays with their respective specific primers (Supplementary Table S4). All the qPCR assays were performed using a 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) using SYBR green chemistry (SuperReal PreMix Plus, Tiangen Biotech Co., Ltd., China). DNA extract (1 ╬╝L) was added to the amplification reaction (20 ╬╝L), which contained 0.3 ╬╝L of each primer, 7.9 ╬╝L of 2├ŚSuper Real PreMix Plus, and 8 ╬╝L of ddH2O. The thermal cycling conditions were as follows: an initial Taq activation step at 95┬░C for 5 min, 40 cycles of denaturation at 95┬░C for 15 s, annealing at 60┬░C for 30 s, followed by an amplicon dissociation stage (95┬░C for 15 s and 60┬░C for 1 min, increasing 0.5┬░C/cycle until 95┬░C was reached). Fluorescence detection was performed at the end of each denaturation and extension step. Following qPCR, the amplicon products were confirmed by agarose gel (1.2%) electrophoresis. To minimize variations, three replicates of each genomic DNA samples were used and a negative control was loaded on each plate run to screen for possible contamination or dimer formation.
The absolute abundance was calculated by multiplying the number of copies per real-time PCR and the number of reactions that can be done with the DNA derived from 1 gram or ml of each sample, while the relative abundance was calculated by dividing the absolute abundance by the corresponding total bacterial abundance in each sample and then multiplying by 1 million.
Ruminal fermentation parameters, bacterial community metrics (Shannon index, even OTUs, abundance-based coverage estimator [ACE], observed species), phylum and genus level abun-dance were performed with PROC T TEST in SAS 9.4 (SAS Institute, Cary, NC, USA) and plotted with Prism 5.0 (GraphPad Software, La Jolla, CA, USA). Abundance of examined selective bacteria species were transformed to ln before analysis. Significance was defined as pŌēż0.05 and trends were defined at a level of 0.05<pŌēż0.10. For comparing the effect on bacterial community, sequences unclassified at the genus level and genus with <0.10% percent relative sequence abundance were not included.
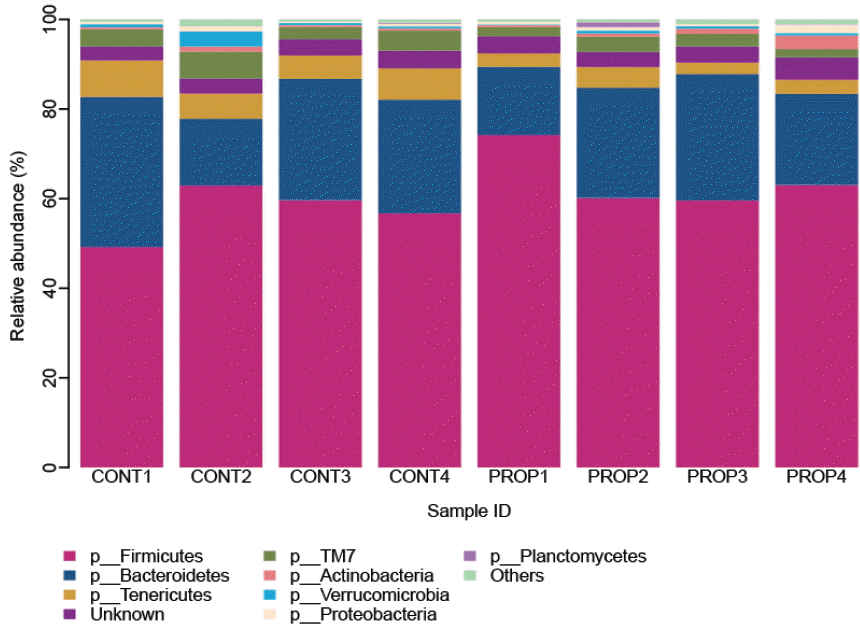
A total of 194,684 reads were analyzed in this study after the stringent quality control. The number of sequence returned into each sample was from 12,942 to 49,092 (Supplementary Table S1). And, a total of 52,873 OTUs were calculated for eight samples at 3% dissimilarity. The average OTUs for CONT and PROP feeding groups were 8,334 and 4,883 per sample, respectively. After normalization to same size of 12,000 reads, richness estimates and diversity indices were developed. Rarefaction analysis, built to assess the sampling saturation, revealed highly diverse bacterial communities and complete sampling for most samples. Only those represented at more than 0.3% of the relative abundance were considered, and overall 8 phyla of bacteria were identified in the rumen samples (Figure 1 and Supplementary Table S2). Most sequences were assigned to two phyla irrespective of diets, averaging the percentages between treatments in Table 2, Firmicutes were highly abundant accounting for 60.68% of total sequence, followed by Bacteroidetes (23.67% of total sequence). And 3.69% sequence unclassified into unknown phyla. Some minor phyla such as Tenericutes, TM7, Actinobacteria, Verrucmicrobia, Proteobacteria, and Planctomycetes accounted for 4.95%, 3.39%, 0.96%, 0.85%, 0.80%, and 0.19% respectively. The Cyanobacteria, SR1, Thermi and Planctomycetes were not present in all samples. The majority of bacteria detected in the Firmicutes phylum belonged to the class Clostridia, which accounted for 46.3% of all rumen bacterial sequences. And Bacteroidia (23.6%) was main class of Bacteroidetes. Majority sequence of Clostridiales order was assigned into Lachnospiraceae (11.35%) followed by Ruminococcaceae (9.81%) and Veillonellaceae (9.39%), while the Bacteroidiales order was mainly occupied by S24ŌĆō7 family (7.01%) followed by Prevotellaceae family. At genus level, only 8 genera were present at more than 0.30% of the relative abundance. The average percentage of the two treatments of the genera Succiniclasticum, Butyrivibrio, Ruminococcus, and Prevotella were 9.43%, 3.74%, 3.46%, and 2.86% respectively and an average of 74.79% sequences represented unclassified at the genus level.
Table 1 shows the effects of propionate supplementation on the rumen fermentation parameters. Results indicated no differences were observed in rumen pH value and NH3-N concentration between CONT and PROP feeding treatments. Total and individual VFA concentrations were not influenced by the feeding treatments. Figure 2 shows the effect of propionate supplementation on the diversity of ruminal bacteria. Shannon index and observed species did not show a difference tendency (p = 0.8275; p = 0.6704) between two treatments. In addition, even OTUs (p = 0.6222) and ACE (p = 0.8068) index of samples were not influenced by propionate supplementation. As shown in Figure 3A, 4,510 OTUs (20.47%) were shared among the two feeding groups, whereas 9,149 OTUs (41.53%) could be detected in PROP group, and 8,372 OTUs (38.00%) were detected in CONT group. For clarity and visualization purposes, the top 50 bacterial OTUs are presented in Thermal double dendrogram. The result demonstrated that all samples from CONT can be grouped with one PROP sample, while the rest of samples from PROP were grouped into another cluster (Figure 3C). Meanwhile, the two treatments could not be obviously separated each other, meaning that the bacterial communities of different treatments were substantially no different. PCoA of overall diversity based on an unweighted UniFrac metric was performed to compare all samples. The dividing line was not obvious and the two principal components covered 33.16% of the variation, the CONT feeding group samples could not be distinguished from the samples of the PROP feeding group (Figure 3B).
To assess the potential of calcium propionate supplementation influence on bacterial communities in the rumen, pyrosequencing data sets from individual groups of bulls were examined. The results are shown in Table 2. The proportion of Firmicutes, the major Gram-positive phylum in the rumen, was not affected in PROP feeding group given propionate supplementation (p = 0.1631). And the proportion of Bacteroidetes, the major Gram-negative phylum, was also similar in the two groups (p = 0.5371). As for the less abundant phyla, nearly all classified phyla had no difference between the feeding treatments (p>0.05), except that Tenericutes (p = 0.0078) was decreased, and TM7 (p = 0.0633) showed a decreasing tendency with the propionate supplementation. Predominant genera did not show a difference between the two feeding groups (p>0.05) (Table 3 and Supplementary Table S3), except YRC22 (p = 0.0120) was decreased with the propionate supplementation.
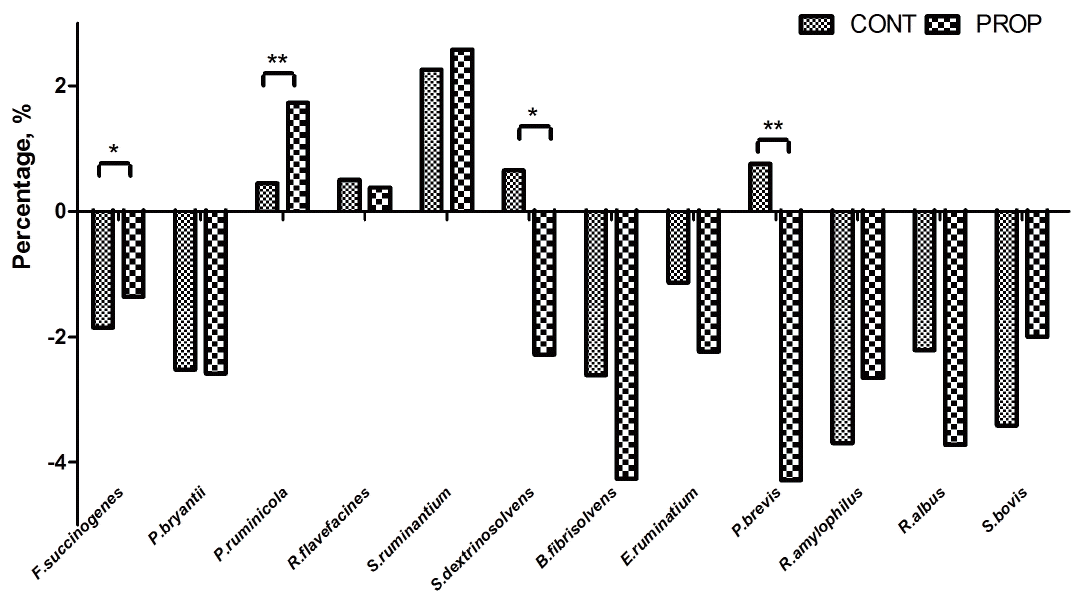
Real-time qPCR analysis of twelve different bacterial targets are shown in Figure 4. Compared with CONT group, the proportion of Fibrobacter succinogenes (p = 0.020) and Prevotella ruminicola (p = 0.009), were found to be increased by propionate supplementation, while that of Succinivibrio dextrinosolvens (p = 0.013) and Prevotella brevis (p = 0.006) were significantly declined. And there was no difference in the proportion of Ruminococcus flavefacines between the two groups. P. ruminicola, and S. ruminantium accounted for a large percentage of the total bacteria in the control treatment (1.56% and 9.61%, respectively) and contributed the least to the propionate supplementation diet (5.67% and 13.18%, respectively). The rest of the examined bacterial species had no difference between the treatments, which accounted for the smallest proportion of the bacterial population (less than 0.32%).
How to improve ruminal fermentation is one of the fundamental questions regarding variability in the composition and activities of the ruminal microbiota. Previous studies also showed that addition of sodium or calcium propionate salt could impact ruminant performance [6,18ŌĆō21]. Supplementing with calcium propionate salts has increased propionate and decreased A:P ratio in the rumen of beef cattle and sheep [5,6,8]. While in our present study, no differences were detected in rumen fermentation parameters between the treatments, including propionate concentration. Another report also showed that propionate salt in a concentrate based lamb diet (65%, dry matter) had no effect on the concentration of propionate in the rumen of finishing lambs consuming mixed rations [9], which was similar to our results. The diet differences between the studies may be the fundamental factor for the variability of propionate concentration. Propionate salt supplementation could probably increase propionate concentration in forage based diets, such as heifers receiving a basal ration of chopped hay [5] or steers consuming rations containing 60% maize stover [6], while there was no effect on a concentrate based diet [9]. In agreement with our results, no differences in ruminal pH were observed in ruminant fed feedlot or other mixed rations with propionate supplementation [8,9]. And there were also no differences detected in pH in steers with a low quality forage based diet [5]. In agreement with our results, no differences in ruminal ammonia concentrations were observed in beef cattle fed feedlot or low quality forage [5,6], which is consistent with the similar bacterial community between the treatments. However, other researchers have reported ruminal ammonia decreased with propionate supplementation [8]. Animal breed variety may be contribute to the difference in ruminal ammonia concentration. More research should be conducted to explore the relationship between ammonia change and breeds with greater numbers of animals.
Bacterial community variations might be linked to the change of physiological parameters in their hosts. It has been reported that supplementation in the diets could influence the diversity and community composition of ruminal microbes [22ŌĆō26]. For comparing the response of the ruminal bacterial community to propionate supplementation, we presently used pyrosequencing of the V3 region of the 16S rRNA gene to create large scale profiles of the bacterial communities. The V3 region was chosen as its length was within the sequencing metrics of the platform, and its relatively consistent amplification and community detection profile. As expected, members of the phylum Firmicutes and Bacteroidetes constituted the major fractions of the total sequencing reads irrespective of the diet, which in agreement with previous findings [22,27ŌĆō30]. While abundance of Tenericutes decreased along with the calcium propionate addition, and TM7 also showed a declining trend. TM7 and Tenericutes have been detected in a wide range of environments including soils, water, activated sludge, and termite guts [31]. It should be noted that microorganisms from all three domains (bacterial and Archaea, ciliate, anaerobic fungi) of life form a complex network in the rumen ecosystem to ferment the feed material ingested by the ruminant. Protozoa and fungi, although far less abundant than the bacteria and archaea in terms of cell numbers, can make up approximately half of the total rumen microbial biomass. Studies are needed to examine the effect of propionate supplementation on the diversity of Archaea, ciliate and anaerobic fungi.
Succiniclasticum is the main propionate-producing genera, and it obtains metabolic energy by converting succinate to propionate. Our result was similar to a previous report [32], in which Succiniclasticum was particularly prevalent when animals were fed a high concentration diet. Succinate utilization to produce propionate would be important in a high concentrate diet, as the fermentation of lactate stabilizes the rumen environment and prevents lactic acid build-up [33]. Genus Prevotella can constitute around half of the total bacterial 16S rRNA gene copy number [17]. While our result showed Prevotella accounting for small proportion, even less than Butyrivibrio and Ruminococcus. One probable explanation is the differences in diet composition fed to the animals or the testing animal species. Another reason is the different molecular method used to assess the microbial community. Our result is similar to one recent report [34], in which Prevotella accounted for 3.66% in the high grain feeding group, which suggests that there may be other bacterial taxa responsible for the fermentation pathways of high concentration diets. Previous study also revealed that high concentration or starch in the diet could decrease the abundance of Prevotella in the rumen [35]. We also noticed that the Firmicutes and Bacteroidetes phyla were under represented at the genus level, which means that there are still a large number of micro-organisms in the rumen that have not been recognized. Due to the large number of unclassified genera and species level taxa in sequencing data bases, pyrosequencing information has limitations in its ability to fully elucidate the complete bionetwork of the rumen ecosystem. Reference databases, such as Greengens, need to be expanded to include more rumen microbial genomes as well as identify a greater number of uncultured species.
R. albus, R. flavefacines, and F. succinogenes are presently recognized as the major cellulolytic bacterial species found in the rumen, and S. ruminantium is the predominant hemicellulose-degrading bacteria. They are all thought to be pivotal in the maintenance of rumen function, because they can convert plant fiber, the major components of plant biomass, to low-molecular-weight carbohydrates and VFAs [36]. Although the populations of F. succinogenes was significantly increased, the cellulolytic bacteria, such as R. albus and R. flavefacines, were unchanged in PROP treatment. Interpretation the effect of propionate feeding on cellulose degradation is a remaining part for further study on the basis of the results. Amylolytic, non-cellulolytic species of rumen bacteria, including P. bryantii, P. ruminicola, S. bovis and R. amylophilus in the present study, hydrolyze starch with the production of intracellular amylase to glucose, maltose, maltotriose, and maltotetraose [37]. Except for the abundance of P. ruminicola that was significantly increased in PROP feeding group, the other amylolytic species level were similar between groups. The increasing tendency with the species in PROP feeding group demonstrated that propionate supplementation probably had a positive effect on degrading of amylum.
This work offers new insights into the effects of calcium propionate addition in finishing diets on rumen microbiota. Pyrosequencing showed that propionate did not change the diversity of the ruminal bacterial community, which provides a basis for designing strategies for the development of feed additives for the manipulation of ruminal microflora as well as guaranteeing improvement in animal production. These results underline the complexity of the rumen ecosystem response to dietary propionate supplementation and the need for further research.
ACKNOWLEDGMENTS
This work was supported by grants from the China Agricultural Research System (CARS-38), the National Natural Science Foundation of China (31372335), and Special Fund for Agro-scientific Research in the Public Interest (201503134). And a scholarship awarded to Z.Z. from the Sino-Dutch Dairy Development Centre supported the tenure of Z.Z. at University of Wageningen where part of this study was done. We gratefully acknowledge Xinzhuang Zhang and Lin Lu for their feeding the animals and technical assistance.
Figure┬Ā2
Bacterial community diversity between the control feeding group (CONT) and calcium propionate supplementation feeding group (PROP). No significant differences in bacterial communities were observed between the two groups. Sequences were normalized to the depth of 12,000 sequences (A) Shannon index. (B) Observed species. (C) Even operational taxonomic units. (D) abundance-based coverage estimator (ACE).

Figure┬Ā3
Differences in OTUs of ruminal bacteria between the control feeding group (CONT) and calcium propionate supplementation feeding group (PROP). (A) Venn diagram showing the number of shared OTU between feeding groups. (B) Principal coordinate analysis (PCoA) of bacterial community structures of the ruminal microbiota of the two groups. The PCoA plots were constructed using the unweighted UniFrac method. Blue squares and red circles represent PROP and CONT feeding group samples, respectively. (C) Thermal double dendrogram of the most abundant bacterial operational taxonomic units (OTU).

Figure┬Ā4
Difference in relative abundance of selected bacterial species between control feeding group (CONT) and calcium propionate supplementation (PROP). Percent data were common log-transformed prior to analysis. Asterisk shows significant differences between groups (** p<0.01, * p<0.05, T test, SAS, Institute, Cary, NC, USA).
Table┬Ā1
Effect of calcium propionate feeding on ruminal fermentation parameters
| Items | CONT | PROP | Equality of variances | Pr > |t|1) | ||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Mean | SD | Mean | SD | |||
| pH | 5.49 | 0.18 | 5.39 | 0.27 | 0.5095 | 0.5690 |
| Ammonia (mmol/L) | 0.94 | 0.09 | 1.03 | 0.22 | 0.1739 | 0.5023 |
| Total VFA (mmol/L) | 108.81 | 5.11 | 108.18 | 5.49 | 0.9098 | 0.8712 |
| Acetate (mmol/L) | 69.54 | 3.60 | 68.35 | 3.22 | 0.8601 | 0.6384 |
| Propionate (mmol/L) | 21.73 | 2.22 | 21.88 | 2.82 | 0.7050 | 0.9361 |
| Butyrate (mmol/L) | 13.05 | 0.91 | 13.40 | 1.05 | 0.8237 | 0.6335 |
| Isobutyrate (mmol/L) | 0.81 | 0.08 | 0.85 | 0.07 | 0.9654 | 0.5299 |
| Valerate (mmol/L) | 1.55 | 0.23 | 1.69 | 0.22 | 0.9618 | 0.4383 |
| Isovalerate (mmol/L) | 2.14 | 0.24 | 2.01 | 0.23 | 0.9405 | 0.4932 |
| Acetate to propionate ratio | 3.21 | 0.17 | 3.18 | 0.03 | 0.0203 | 0.6903 |
Table┬Ā2
Effect of calcium propionate feeding on the difference in most abundant phyla (as a percentage of the total sequences) in the rumen bacterial community
| Taxon | CONT | PROP | Equality of variances | Pr>|t|1) | ||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Mean | SD | Mean | SD | |||
| Firmicutes | 57.11 | 5.85 | 64.24 | 6.81 | 0.8097 | 0.1631 |
| Bacteroidetes | 25.23 | 7.72 | 22.10 | 5.61 | 0.6141 | 0.5371 |
| Tenericutes | 6.54 | 1.34 | 3.37 | 0.91 | 0.5384 | 0.0078 |
| TM7 | 4.27 | 1.35 | 2.52 | 0.73 | 0.3358 | 0.0633 |
| Verrucomicrobia | 1.25 | 1.46 | 0.46 | 0.20 | 0.0088 | 0.3625 |
| Proteobacteria | 0.66 | 0.33 | 0.94 | 0.63 | 0.3138 | 0.4468 |
| Actinobacteria | 0.61 | 0.33 | 1.32 | 1.16 | 0.0670 | 0.2823 |
| SR1 | 0.08 | 0.06 | 0.08 | 0.01 | 0.0050 | 0.8980 |
| Chloroflexi | 0.16 | 0.05 | 0.17 | 0.08 | 0.5065 | 0.7991 |
| Cyanobacteria | 0.11 | 0.04 | 0.06 | 0.03 | 0.6780 | 0.1185 |
| [Thermi] | 0.06 | 0.03 | 0.06 | 0.03 | 0.7737 | 0.9184 |
| Planctomycetes | 0.07 | 0.11 | 0.30 | 0.51 | 0.0332 | 0.4451 |
| Spirochaetes | 0.02 | 0.01 | 0.03 | 0.01 | 0.6645 | 0.2528 |
| WPS-2 | 0.01 | 0.00 | 0.01 | 0.01 | 0.3142 | 0.3903 |
| Elusimicrobia | 0.00 | 0.00 | 0.00 | 0.00 | <0.0001 | 0.3910 |
| Fibrobacteres | 0.01 | 0.01 | 0.01 | 0.01 | 0.7998 | 0.7049 |
| Lentisphaerae | 0.00 | 0.00 | 0.01 | 0.02 | 0.0821 | 0.4772 |
| Synergistetes | 0.01 | 0.01 | 0.02 | 0.01 | 1.0000 | 0.2070 |
| Unclassified phyla | 3.46 | 0.35 | 3.92 | 0.73 | 0.2676 | 0.3015 |
Table┬Ā3
Effect of calcium propionate feeding on the difference in most abundant genus (as a percentage of the total sequences) in the rumen bacterial community
| Taxon | CONT | PROP | Equality of variances | Pr>|t|1) | ||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Mean | SD | Mean | SD | |||
| Butyrivibrio | 3.94 | 0.91 | 3.54 | 0.71 | 0.6913 | 0.5098 |
| Succiniclasticum | 11.35 | 3.70 | 7.52 | 5.33 | 0.5625 | 0.2817 |
| Ruminococcus | 3.35 | 1.12 | 3.56 | 1.72 | 0.5043 | 0.8488 |
| Prevotella | 2.70 | 0.67 | 3.01 | 0.66 | 0.9814 | 0.5370 |
| Mogibacterium | 0.65 | 0.56 | 0.54 | 0.30 | 0.3262 | 0.7332 |
| p-75-a5 | 0.90 | 0.41 | 0.46 | 0.24 | 0.3984 | 0.1143 |
| Bifidobacterium | 0.19 | 0.30 | 0.46 | 0.49 | 0.4427 | 0.3853 |
| Anaerostipes | 0.75 | 0.49 | 0.36 | 0.17 | 0.1210 | 0.1841 |
| Coprococcus | 0.36 | 0.12 | 0.38 | 0.08 | 0.4954 | 0.8016 |
| YRC22 | 0.18 | 0.04 | 0.29 | 0.05 | 0.9044 | 0.0120 |
| CF231 | 0.14 | 0.03 | 0.25 | 0.12 | 0.0531 | 0.1370 |
| L7A_E11 | 0.20 | 0.08 | 0.18 | 0.04 | 0.2132 | 0.6690 |
| Anaerovibrio | 0.43 | 0.17 | 0.38 | 0.27 | 0.4603 | 0.7922 |
| Blautia | 0.14 | 0.05 | 0.14 | 0.07 | 0.6023 | 1.0000 |
| Bulleidia | 0.10 | 0.05 | 0.18 | 0.06 | 0.7080 | 0.0774 |
| SHD-231 | 0.16 | 0.05 | 0.17 | 0.08 | 0.5065 | 0.7991 |
| Clostridium | 0.10 | 0.04 | 0.14 | 0.03 | 0.5205 | 0.1289 |
| Unclassified genera | 72.77 | 4.83 | 76.80 | 7.92 | 0.4374 | 0.4180 |
REFERENCES
1. Nakamura I, Ogimoto K, Imai S, Nakamura M. Production of lactic-acid isomers and change of microbial features in the rumen of feedlot cattle. J Anim Physiol Anim Nutr 1989; 61:139ŌĆō44.

2. Aiello RJ, Armentano LE, Bertics SJ, Murphy AT. Volatile fatty acid uptake and propionate metabolism in ruminant hepatocytes. J Dairy Sci 1989; 72:942ŌĆō9.


3. Seal CJ, Reynolds CK. Nutritional implications of gastrointestinal and liver metabolism in ruminants. Nutr Res Rev 1993; 6:185ŌĆō208.


4. Rigout S, Hurtaud C, Lemosquet S, Bach A, Rulquin H. Lactational effect of propionic acid and duodenal glucose in cows. J Dairy Sci 2003; 86:243ŌĆō53.


5. Sanchez PH, Tracey LN, Browne-Silva J, Lodge-Ivey SL. Propionibacterium acidipropionici P169 and glucogenic precursors improve rumen fermentation of low-quality forage in beef cattle. J Anim Sci 2014; 92:1738ŌĆō46.


6. Liu Q, Wang C, Guo G, et al. Effects of calcium propionate on rumen fermentation, urinary excretion of purine derivatives and feed digestibility in steers. J Agric Sci 2009; 147:201ŌĆō9.

7. McNamara JP, Valdez F. Adipose tissue metabolism and production responses to calcium propionate and chromium propionate. J Dairy Sci 2005; 88:2498ŌĆō507.


8. Moloney AP. Growth and carcass composition in sheep offered isoenergetic rations which resulted in different concentrations of ruminal metabolites. Livest Prod Sci 1998; 56:157ŌĆō64.

9. Lee-Rangel HA, Mendoza GD, Gonz├Īlez SS. Effect of calcium propionate and sorghum level on lamb performance. Anim Feed Sci Technol 2012; 177:237ŌĆō41.

10. Miettinen H, Huhtanen P. Effects of the ratio of ruminal propionate to butyrate on milk yield and blood metabolites in dairy cows. J Dairy Sci 1996; 79:851ŌĆō61.


11. Hurtaud C, Rulquin H, Verite R. Effects of level and type of energy source (volatile fatty acids or glucose) on milk yield, composition and coagulating properties in dairy cows. Reprod Nutr Dev 1998; 38:315ŌĆō30.


12. Broderick GA, Kang JH. Automated simultaneous determination of ammonia and total amino acids in ruminal fluid and in vitro media. J Dairy Sci 1980; 63:64ŌĆō75.


13. Erwin ES, Marco GJ, Emery EM. Volatile fatty acid analyses of blood and rumen fluid by gas chromatography. J Dairy Sci 1961; 44:1768ŌĆō71.

14. Ding G, Chang Y, Zhao L, et al. Effect of Saccharomyces cerevisiae on alfalfa nutrient degradation characteristics and rumen microbial populations of steers fed diets with different concentrate-to-forage ratios. J Anim Sci Biotechnol 2014; 5:24



15. Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010; 7:335ŌĆō6.



16. Weimer PJ, Stevenson DM, Mertens DR, Thomas EE. Effect of monensin feeding and withdrawal on populations of individual bacterial species in the rumen of lactating dairy cows fed high-starch rations. Appl Microbiol Biotecnol 2008; 80:135ŌĆō45.

17. Stevenson DM, Weimer PJ. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl Microbiol Biotech 2007; 75:165ŌĆō74.

18. Sheperd AC, Combs DK. Long-term effects of acetate and propionate on voluntary feed intake by midlactation cows. J Dairy Sci 1998; 81:2240ŌĆō50.


19. Beiranvand H, Ghorbani GR, Khorvash M, et al. Interactions of alfalfa hay and sodium propionate on dairy calf performance and rumen development. J Dairy Sci 2014; 97:2270ŌĆō80.


20. Pehrson B, Svensson C, Jonsson M. A comparative study of the effectiveness of calcium propionate and calcium chloride for the prevention of parturient paresis in dairy cows. J Dairy Sci 1998; 81:2011ŌĆō6.


21. Ferreira LS, Bittar CM. Performance and plasma metabolites of dairy calves fed starter containing sodium butyrate, calcium propionate or sodium monensin. Animal 2011; 5:239ŌĆō45.


22. Kim M, Eastridge ML, Yu Z. Investigation of ruminal bacterial diversity in dairy cattle fed supplementary monensin alone and in combination with fat, using pyrosequencing analysis. Can J Microbiol 2014; 60:65ŌĆō71.


23. Zhou Z, Yu Z, Meng Q. Effects of nitrate on methane production, fermentation, and microbial populations in in vitro ruminal cultures. Bioresour Technol 2012; 103:173ŌĆō9.


24. Mohammed R, Zhou M, Koenig KM, Beauchemin KA, Guan LL. Evaluation of rumen methanogen diversity in cattle fed diets containing dry corn distillers grains and condensed tannins using PCR-DGGE and qRT-PCR analyses. Anim Feed Sci Technol 2011; 166ŌĆō67:122ŌĆō31.

25. Pinloche E, McEwan N, Marden JP, et al. The effects of a probiotic yeast on the bacterial diversity and population structure in the rumen of cattle. PLoS One 2013; 8:e67824



26. Zhou Z, Meng Q, Yu Z. Effects of methanogenic inhibitors on methane production and abundances of methanogens and cellulolytic bacteria in in vitro ruminal cultures. Appl Environ Microbiol 2011; 77:2634ŌĆō9.



27. Thoetkiattikul H, Mhuantong W, Laothanachareon T, et al. Comparative analysis of microbial profiles in cow rumen fed with different dietary fiber by tagged 16S rRNA gene pyrosequencing. Curr Microbiol 2013; 67:130ŌĆō7.


28. Ozutsumi Y, Tajima K, Takenaka A, Itabashi H. The effect of protozoa on the composition of rumen bacteria in cattle using 16S rRNA gene clone libraries. Biosci Biotech Bioch 2005; 69:499ŌĆō506.

29. Kim M, Felix TL, Loerch SC, Yu Z. Effect of haylage and monensin supplementation on ruminal bacterial communities of feedlot cattle. Curr Microbiol 2014; 69:169ŌĆō75.


30. Pitta DW, Kumar S, Veiccharelli B, et al. Bacterial diversity associated with feeding dry forage at different dietary concentrations in the rumen contents of Mehshana buffalo (Bubalus bubalis) using 16S pyrotags. Anaerobe 2014; 25:31ŌĆō41.


31. Kurogi T, Linh NTT, Kuroki T, Yamada T, Hiraishi A. Culture-independent detection of ŌĆ£TM7ŌĆØ bacteria in a streptomycin-resistant acidophilic nitrifying process. Aip Conf Proc 2014; 1585:53ŌĆō8.

32. Yang SL, Ma SC, Chen J, et al. Bacterial diversity in the rumen of Gayals (Bos frontalis), Swamp buffaloes (Bubalus bubalis) and Holstein cow as revealed by cloned 16S rRNA gene sequences. Mol Biol Rep 2010; 37:2063ŌĆō73.


33. Stewart DJ. Biochemical and biological studies on the lipopolysaccharide of Bacteroides nodosus. Res Vet Sci 1977; 23:319ŌĆō25.


34. Liu JH, Bian GR, Zhu WY, Mao SY. High-grain feeding causes strong shifts in ruminal epithelial bacterial community and expression of Toll-like receptor genes in goats. Front Microbiol 2015; 6:Article 167



35. Zened A, Combes S, Cauquil L, et al. Microbial ecology of the rumen evaluated by 454 GS FLX pyrosequencing is affected by starch and oil supplementation of diets. FEMS Microbiol Ecol 2013; 83:504ŌĆō14.


- TOOLS
-
METRICS

- Related articles
-
Effect of fibrous diets on chemical composition and odours from pig slurry2018 November;31(11)





 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print




