 |
 |
| Anim Biosci > Volume 29(3); 2016 > Article |
|
Abstract
Porcine beta-defensin-1 (PBD-1) gene plays an important role in the innate immunity of pigs. The peptide encoded by this gene is an antimicrobial peptide that has direct activity against a wide range of microbes. This peptide is involved in the co-creation of an antimicrobial barrier in the oral cavity of pigs. The objective of the present study was to detect polymorphisms, if any, in exon-1 and exon-2 regions of PBD-1 gene in Large White Yorkshire (LWY) and native Ankamali pigs of Kerala, India. Blood samples were collected from 100 pigs and genomic DNA was isolated using phenol chloroform method. The quantity of DNA was assessed in a spectrophotometer and quality by gel electrophoresis. Exon-1 and exon-2 regions of PBD-1 gene were amplified by polymerase chain reaction (PCR) and the products were subjected to single strand conformation polymorphism (SSCP) analysis. Subsequent silver staining of the polyacrylamide gels revealed three unique SSCP banding patterns in each of the two exons. The presence of single nucleotide polymorphisms (SNPs) was confirmed by nucleotide sequencing of the PCR products. A novel SNP was found in the 5′-UTR region of exon-1 and a SNP was detected in the mature peptide coding region of exon-2. In exon-1, the pooled population frequencies of GG, GT, and TT genotypes were 0.67, 0.30, and 0.03, respectively. GG genotype was predominant in both the breeds whereas TT genotype was not detected in LWY breed. Similarly, in exon-2, the pooled population frequencies of AA, AG, and GG genotypes were 0.50, 0.27, and 0.23, respectively. AA genotype was predominant in LWY pigs whereas GG genotype was predominant in native pigs. These results suggest that there exists a considerable genetic variation at PBD-1 locus and further association studies may help in development of a PCR based genotyping test to select pigs with better immunity.
Disease occurrence is a major threat to any livestock farming and pig production is no exception. Diseases cause high morbidity and mortality resulting in huge loss to farmers. Control of diseases has traditionally been accomplished through the use of sanitation, medication, vaccination and elimination of sick pigs but, these methods have often been proved to be inadequate. Selection for disease resistance employing molecular markers would help in evolving a population with greater immunity. Modern techniques like polymerase chain reaction-single strand conformation polymorphism (PCR-SSCP) are helpful in identifying such molecular markers.
Antimicrobial peptides (AMPs) are often referred to as natural antibiotics due to their antimicrobial action against a wide range of microbes. AMPs are polypeptides made up of less than 100 amino acid residues (Ganz, 2003). Based on the net charge present, AMPs are broadly classified into anionic and cationic peptides (Hancock, 1997). Defensins are a subclass of cationic AMPs with broad spectrum antimicrobial activity against various bacteria, fungi and viruses. They possess six to eight highly conserved cysteine residues that form intramolecular disulfide bonds based on which, three families of defensins are defined viz. α-, β- and θ-defensins (Lai and Gallo, 2009).
The porcine beta-defensin-1 (PBD-1) gene first reported by Zhang et al. (1999) encodes a peptide that was found to be active against Escherichia coli, Salmonella typhimurium, Listeria monocytogenes, Staphylococcus aureus, and Candida albicans (Shi et al., 1999; Jiang et al., 2006; Li et al., 2013). The β-defensin genes in human, cattle and poultry have been found to be associated with several disease conditions (Bagnicka et al., 2007; Hasenstein and Lamont, 2007; Baroncelli et al., 2008). Similar studies in pigs would help in realising the potential of β-defensins such as PBD-1.
Indian native pigs have unique qualities such as disease resistance, adaptation to the local environment and possession of lean fat (De et al., 2013). Studies on genes responsible for immunity would aid in selection for disease resistance. Therefore, the present investigation was undertaken to assess the genetic variability, if any, at exon-1 and -2 regions of PBD-1 gene in native and exotic populations. In addition, to sequence this portion from the native breed and compare it with the published sequences available in the NCBI database.
The polymorphism study was conducted on a total of 100 animals belonging to the species Sus scrofa (domestic pig). Blood samples were collected from 50 Large White Yorkshire (LWY) and 50 Ankamali pigs reared at Centre for Pig Production and Research, Mannuthy, Thrissur, Kerala, India.
Five millilitre of blood was collected from the ear vein of each animal into a sterile 15 mL polypropylene centrifuge tube containing ethylene diamine tetra acetic acid (EDTA) as anticoagulant (1 mg/mL of blood). The tubes were tightly capped and kept in an ice box. Samples were taken to the molecular biology laboratory of Centre for Advanced Studies in Animal Genetics and Breeding and stored at −20°C until isolation of DNA. Genomic DNA was isolated from whole blood using standard phenol chloroform method (Sambrook and Russell, 2001) with necessary modifications.
A 143 bp fragment (partial promoter and complete exon-1) and a 322 bp fragment (partial intron and complete exon-2) of PBD-1 gene were amplified using two sets of primers (Table 1). PCR was carried out in a thermal cycler (Bio-Rad, Hercules, CA, USA) using 0.2 mL PCR tubes. Each 25 μL volume reaction contained 1× Taq polymerase buffer, 1.5 mM MgCl2, 0.2 mM of dNTP mixture, 10 pM/μL each of forward and reverse primers, 50 to 100 ng of template DNA, 0.75 units of Taq polymerase and nuclease free water to make up the volume.
The PCR conditions followed were initial denaturation for 3 min at 94°C, followed by 35 cycles of 30 s at 95°C, 20 s at 58.8°C, 30 s at 72°C and final extension at 72°C for 8 min. The amplified PCR products were resolved in 2% (w/v) agarose gel in 1× Tris Borate EDTA (TBE). Five microlitre of PCR product mixed with 1 μL of 6× DNA gel loading dye were loaded in the well. A 50 bp DNA marker was used to ascertain the size of the amplified products. Electrophoresis was performed at 80 V for 10 minutes followed by 65 V for 30 minutes. The gels were visualized under UV light and recorded in a gel documentation system.
Single strand conformation polymorphism (SSCP) was performed using PCR products of exon-1 and exon-2 regions of PBD-1 gene. The PCR products were mixed with SSCP loading dye (95% Formamide dye) in 1:2 ratio (20 μL sample with 40 μL dye), denatured at 95°C for 10 minutes in a thermal cycler and snap chilled on ice for three minutes. The single stranded DNA samples were resolved in a 12% polyacrylamide gel. The composition of polyacrylamide gel was 30% acrylamide/Bis-acrylamide (29:1) 12 mL, 1×TBE 3 mL, Tetramethyl-ethylenediamine (TEMED) 30 μL, 10% ammonium per sulphate (APS) 150 μL, and nuclease free water 14.82 mL.
The SSCP-PAGE was performed without adding glycerol as banding patterns were not clear in gels containing 5% (v/v) glycerol. Electrophoresis was performed at 110 V for 143 bp fragment and at 200 V for 322 bp fragment for 16 hours at 12°C. The SSCP bands were visualized by silver staining following the protocol of Bassam et al. (1991) with certain modifications. The protocol used in the present study had several advantages. Fixing the gels with 10 per cent ethanol instead of 10 per cent acetic acid reduced the time required for fixation and addition of the pre-treatment step with 1.5 per cent nitric acid hardened the gel enabling its easy handling.
Representative PCR products showing different SSCP banding patterns were selected and sequenced using respective forward and reverse primers to detect the polymorphisms, if any, at nucleotide level. Sequencing was performed by automated sequencer (ABI prism, Applied Biosystems, Foster City, CA, USA) using Sanger’s dideoxy chain termination method (Sanger et al., 1977) at SciGenom Labs Pvt. Ltd., Cochin, India. The sequences of different designated alleles of the gene were analysed using the ‘MegAlign’ tool of Lasergene Software (DNASTAR, Madison, WI, USA) to generate sequence alignment reports, sequence distances and phylogenetic trees.
The genotypes were detected by observing the SSCP banding pattern of each sample in the gels which was further confirmed by sequencing. The number of individuals belonging to different genotypes was recorded by direct counting (Falconer and Mackay, 1996) and the frequencies were calculated using following formulae:
where,
In order to ascertain the polymorphism in 143 bp fragment (partial promoter and complete exon-1) (Figure 1a), the PCR product was subjected to SSCP-PAGE. Silver staining revealed three unique SSCP banding patterns which were identified as GG, GT, and TT (Figure 1b). Sequencing results confirmed the presence of a novel SNP with G to T transversion in the 5′ UTR of PBD-1 gene (Figure 1c). The two alleles viz. PBD1-G allele and PBD1-T allele of exon-1 were submitted to the NCBI database and are available with the accession numbers KP862536 and KP862537, respectively. Samples with corresponding banding patterns were designated as GG, GT, and TT genotypes. Studies in human HBD-1 gene have also reported SNPs in the 5′ UTR (Jurevic et al., 2003; Baroncelli et al., 2008; Segat et al., 2014).
The SSCP analysis of 322 bp fragment (partial intron and complete exon-2) (Figure 2a) also revealed three unique SSCP banding patterns identified as AA, AG, and GG (Figure 2b). Sequencing results confirmed the presence of a SNP with A to G transition in the coding region of PBD-1 (Figure 2c). The two alleles viz. PBD1-A allele and PBD1-G allele of exon-2 were given accession numbers KP862536 and KP862537, respectively by the NCBI. Three genotypes formed were designated as AA, AG, and GG. This SNP was previously reported by Choi et al. (2012) using PCR-restriction fragment length polymorphism with restriction enzyme BstNI. Although the SNP was within the coding region, it resulted in a synonymous substitution to the amino acid glutamine.
The genotypic and allelic frequencies for exon-1 and exon-2 based on PCR-SSCP pattern are presented in Table 2. In exon-1, the GG genotype showed high frequency in both the breeds whereas the TT genotype was not detected in LWY breed and also the G allele was found to be predominant in both the breeds. In exon-2, AA genotype was predominant in LWY pigs whereas the GG genotype was predominant in native pigs. Consequently, the G allele was predominant in Ankamali pigs whereas the A allele was found to be predominant in LWY pigs. Choi et al. (2012) had reported a frequency of 0.18 for the minor allele at this locus.
The sequences of same region were subjected to multiple sequence alignment using MegAlign tool. The 143 bp sequences obtained from G and T alleles of Ankamali pig were compared with the sequence of other pig (NCBI accession No. AF132038) available in the NCBI database (Figure 3a). The designated G allele showed 100 per cent homology with the sequence of other pig whereas 99.3 per cent similarity with designated T allele. The nucleotide sequence of designated G allele showed nucleotide ‘G’ at position 63 whereas it was nucleotide ‘T’ in the designated T allele as shown in the Figure 3a. This SNP in the 143 bp product was situated at −23 (minus 23) position in the 5′UTR of PBD-1 gene.
The 322 bp sequences obtained from A and G alleles of Ankamali pig were compared (Figure 3b) with the sequences of the same region of other pig (NCBI accession No. AF132038), Ovis aries sheep beta defensin-1 (SBD-1) gene (NCBI accession No. U75250) and Bos taurus tracheal antimicrobial peptide (TAP) gene (NCBI accession No. L13373). The designated A allele showed 100 per cent similarity with the sequence of other pig whereas it was 99.7 per cent with the designated G allele. The per cent identity of designated A allele with sheep and cattle β-defensins was 74.2 and 73.0 respectively. The nucleotide sequence of designated A allele showed nucleotide ‘A’ at position 188 whereas it was nucleotide ‘G’ in the designated G allele as shown in the Figure 3b. This SNP found in the 322 bp product was situated at position 171 from translation start codon in PBD-1 gene.
The phylogenetic tree from 143 bp fragment revealed close evolutionary relationship between the designated G and T alleles and other pig (NCBI accession No. AF132038). It was evident from the common cluster formed by these sequences (Figure 4a). This was expected as they had a high per cent similarity in their sequences. The phylogenetic tree for 322 bp showed two major branches at the primary node for pig and ruminants. The designated A and G alleles as well as the other pig (NCBI accession No. AF132038) formed a common cluster indicating their close relationship in the evolution. Two unique branches were formed for sheep and cattle in the node for ruminants (Figure 4b). It may be inferred from above results that the PBD-1 gene was quite different from ruminant species in terms of structure, organisation and composition but had no breed differences.
PBD-1 gene plays an important role in the innate immunity of pigs. The peptide encoded by this gene co-creates an antimicrobial barrier in the oral cavity of pigs as it has direct activity against a wide range of microbes. Polymorphism studies of this gene revealed two SNPs including a novel SNP at exon-1 in both native and exotic breeds, indicating a considerable genetic variation at PBD-1 locus which may aid in the marker assisted selection for disease resistance traits. The silver staining protocol used in the present study had several advantages which might be useful to other researchers. The nucleotide sequence analysis indicated that the PBD-1 gene was quite different from other species in terms of structure, organisation and composition. However, there was complete homology between the sequence of native pigs and those available in the NCBI database.
ACKNOWLEDGMENTS
Authors are grateful to Kerala Veterinary and Animal Sciences University for providing the facilities for the conduct of the research.
Figure 1
PCR-SSCP analysis of exon 1 of porcine beta-defensin-1 (PBD-1) gene. (a) PCR amplification of 143 bp fragment of exon-1 of PBD-1 gene. Lane M: 50 bp DNA marker, Lane 1 to 4: 143 bp PCR product. (b) SSCP banding pattern of 143 bp fragment of exon-1 of PBD-1 gene. (c) Sanger sequencing analysis showing a novel SNP with G to T transversion in exon-1 of PBD-1 gene. PCR-SSCP, polymerase chain reaction-single strand conformation polymorphism; SNP, single nucleotide polymorphism.
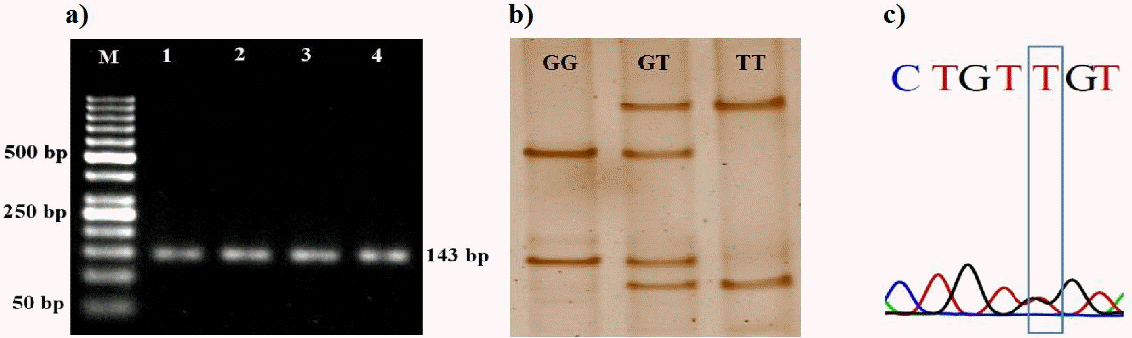
Figure 2
PCR-SSCP analysis of exon 2 of porcine beta-defensin-1 (PBD-1) gene (a) PCR amplification of 322 bp fragment of exon-2 of PBD-1 gene. Lane M: 50 bp DNA marker, Lane 1 to 4: 322 bp PCR product. (b) SSCP banding pattern of 322 bp fragment of exon-2 of PBD-1 gene. (c) Sanger sequencing analysis showing A to G transition in exon-2 of PBD-1 gene. PCR-SSCP, polymerase chain reaction-single strand conformation polymorphism.
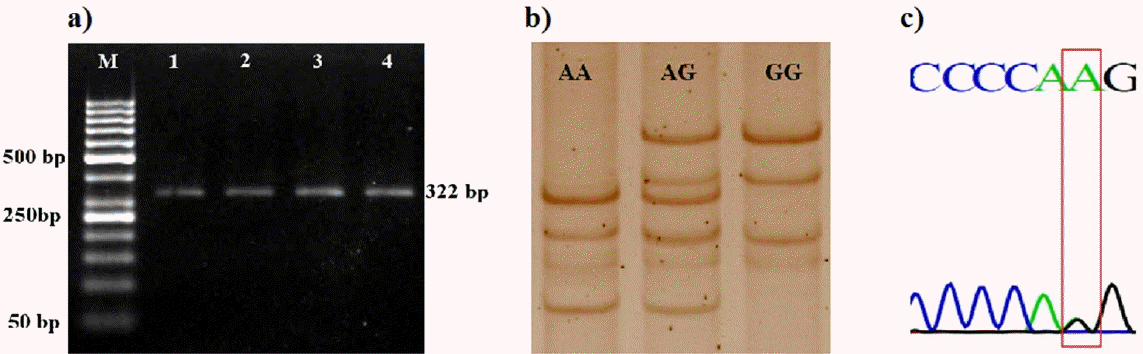
Figure 3
Nucleotide sequences alignment of porcine beta-defensin-1 (PBD-1) gene. (a) Multiple sequence alignment of nucleotide sequences of partial promoter and complete exon-1 of PBD-1 gene in pig under study and other pig (NCBI accession No. AF132038). Shaded portion indicate residues that differ from the consensus. (b) Multiple sequence alignment of nucleotide sequences of partial intron and complete exon-2 of PBD-1 gene in pig under study and other pig (NCBI accession No. AF132038), sheep beta defensin-1 (SBD-1) gene (NCBI accession No. U75250) and bovine tracheal antimicrobial peptide (TAP) gene (NCBI accession No. L13373). Shaded portion indicate residues that differ from the consensus.
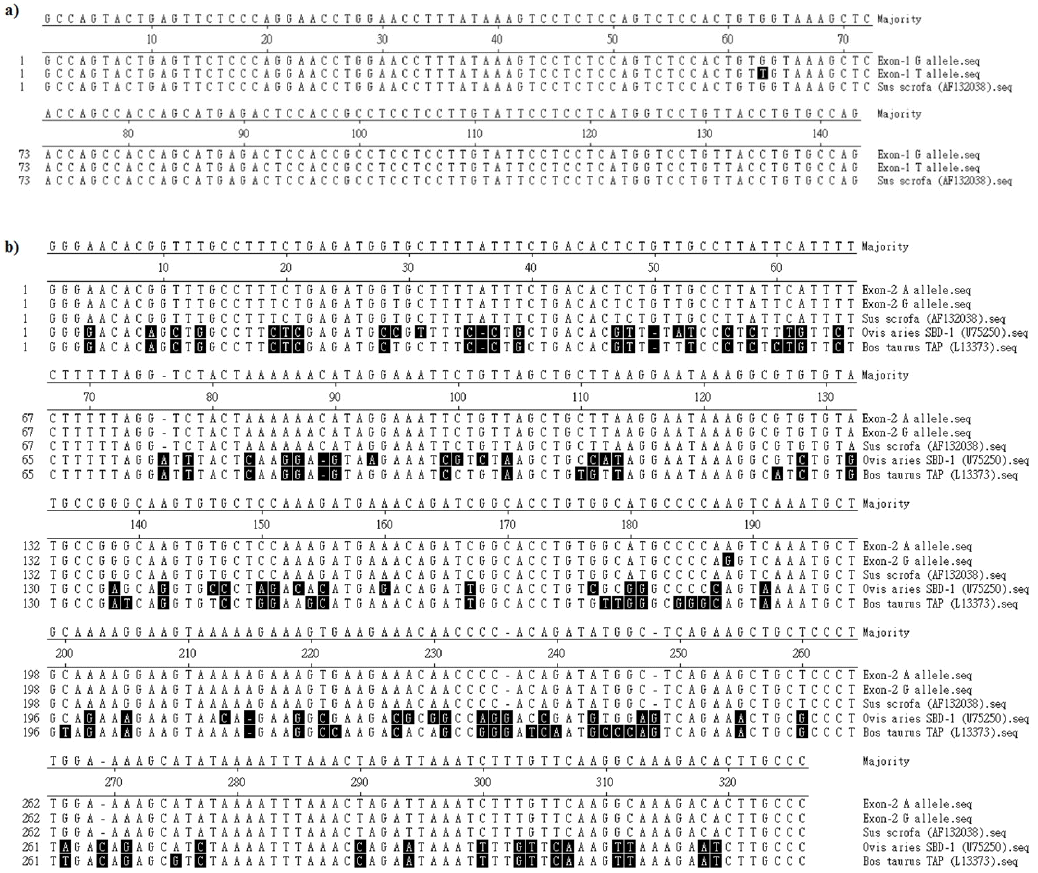
Figure 4
Phylogenetic analysis of porcine beta-defensin-1 (PBD-1) gene. (a) Phylogenetic tree on the basis of nucleotide sequences of partial promoter and complete exon-1 of PBD-1 gene of pig under study with that of other pig (NCBI accession No. AF132038). (b) Phylogenetic tree on the basis of nucleotide sequences of partial intron and complete exon-2 of PBD-1 gene of pig under study with that of other pig (NCBI accession No. AF132038), sheep beta defensin-1 (SBD-1) gene (NCBI accession No. U75250) and bovine tracheal antimicrobial peptide (TAP) gene (NCBI accession No. L13373).
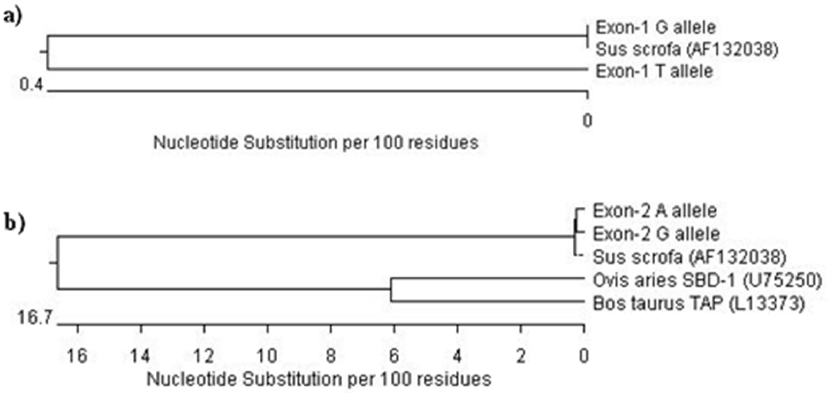
Table 1
Properties and sequences of designed primers for PBD-1 gene exons
Table 2
Genotype and allele frequencies of PBD-1 gene exons1
REFERENCES
Bassam BJ, Caetano-Anollès G, Gresshoff PM. 1991. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 196:80–83.


Bagnicka E, Strzałkowska N, Flisikowski K, Szreder T, Jóźwik A, Prusak B, Krzyżewski J, Zwierzchowski L. 2007. The polymorphism in the β4-defensin gene and its assotiation with production and somatic cell count in Holstein-Friesian cows. J Anim Breed Genet 124:150–156.


Baroncelli S, Ricci E, Andreotti M, Guidotti G, Germano P, Marazzi MC, Vella S, Palombi L, De Rossi A, Giuliano M. 2008. Single-nucleotide polymorphisms in human beta-defensin-1 gene in Mozambican HIV-1-infected women and correlation with virologic parameters. AIDS 22:1515–1517.


Choi MK, Le MT, Nguyen DT, Choi H, Kim W, Kim JH, Chun J, Hyeon J, Seo K, Park C. 2012. Genome-level identification, gene expression, and comparitive analysis of porcine β-defensin genes. BMC Genetics 13:98



De AK, Jeyakumar S, Kundu A, Kundu MS, Sunder J, Ramachandran M. 2013. Genetic characterization of Andaman Desi pig, an indigenous pig germplasm of Andaman and Nicobar group of islands, India by microsatellite markers. Vet Wld 6:750–753.

Falconer DS, Mackay TFC. 1996. Introduction to Quantitative Genetics. 4th EdLongmans Green; Essex, UK:
Hasenstein JR, Lamont SJ. 2007. Chicken gallinacin gene cluster associated with Salmonella response in advanced intercross line. Avian Dis 51:561–567.


Jiang LH, Lu HR, Huang DX, Yi JB, Li LY, Lin F. 2006. Expression of porcine beta-defensin-1 gene in Pichia pastoris. Sheng Wu Gong Cheng Xue Bao 22:1036–1039.

Jurevic RJ, Bai M, Chadwick RB, White TC, Dale BA. 2003. Single-nucleotide polymorphisms (SNPs) in human beta-defensin-1: high-throughput SNP assays and association with Candida carriage in type I diabetics and nondiabetic controls. J Clin Microbiol 41:90–96.



Lai Y, Gallo RL. 2009. AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol 30:131–141.



Li C, Xu T, Chen R, Huang X, Zhao Y, Bao Y, Zhao W, Zheng Z. 2013. Cloning, expression and characterization of antimicrobial porcine β-defensin-1 in Escherichia coli. Protein Expr Purif 88:47–53.


Sambrook J, Russell DW. 2001. Molecular Cloning: A Laboratory Manual. 3rd EdCold Spring Harbor Laboratory Press; New York, NY, USA:
Sanger F, Nicklen S, Coulson AR. 1977. DNA sequencing with chain-terminating inhibitors. Proc Nati Acad Sci USA 74:5463–5467.

Segat L, Zupin L, Moura RR, Coelho AVC, Chagas BS, de Freitas AC, Crovella S. 2014. DEFB1 polymorphisms are involved in susceptibility to human papillomavirus infection in Brazilian gynaecological patients. Mem Inst Oswaldo Cruz 109:918–922.



- TOOLS
-
METRICS

- Related articles
-
Y-Single Nucleotide Polymorphisms Diversity in Chinese Indigenous Horse2015 August;28(8)





 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print




