Targeted Editing of Myostatin Gene in Sheep by Transcription Activator-like Effector Nucleases
Article information
Abstract
Myostatin (MSTN) is a secreted growth factor expressed in skeletal muscle and adipose tissue that negatively regulates skeletal muscle mass. Gene knockout of MSTN can result in increasing muscle mass in sheep. The objectives were to investigate whether myostatin gene can be edited in sheep by transcription activator-like effector nucleases (TALENs) in tandem with single-stranded DNA oligonucleotides (ssODNs). We designed a pair of TALENs to target a highly conserved sequence in the coding region of the sheep MSTN gene. The activity of the TALENs was verified by using luciferase single-strand annealing reporter assay in HEK 293T cell line. Co-transfection of TALENs and ssODNs oligonucleotides induced precise gene editing of myostatin gene in sheep primary fibroblasts. MSTN gene-edited cells were successfully used as nuclear donors for generating cloned embryos. TALENs combined with ssDNA oligonucleotides provide a useful approach for precise gene modification in livestock animals.
INTRODUCTION
Myostatin (MSTN) is a transforming growth factor (TGF)-β superfamily member that plays an important inhibitory role during muscle development (McPherron et al., 1997). Previous studies reported that knockout of MSTN in mice resulted in widespread increase of skeletal muscle mass (McPherron et al., 1997). Injection of MSTN dsRNA in zebrafish embryos resulted in muscle hyperplasia and hypertrophy (Acosta et al., 2005), and mice expressing higher levels of myostatin inhibitors increased skeletal muscle mass (Hu et al., 2010). In line with these findings, several natural mutations (including point and deletion mutations) also leads to increased muscle mass in Piedmontese, Belgian Blue bovine, sheep and dog (Kambadur et al., 1997; Clop et al., 2006; Mosher et al., 2007). These findings suggest that growth performance of normal sheep could be improved by disrupting MSTN function using gene knockout technology.
Traditional gene knockout approach has been limited to mouse embryonic stem cells and not readily adaptable for most other cell types because of the extremely low targeting frequency. The low efficiency of gene targeting in somatic cells is the main barrier for gene knockout in large animals. Recently, transcription activator-like effector nucleases (TALENs) and zinc finger nucleases (ZFNs) technology have emerged as a powerful tool for genome editing (Boch et al., 2009; Moscou et al., 2009). Custom-designed TALENs have been reported to induce indels frequencies between 2% and 55% of targeted chromosomes (Huang et al., 2011; Reyon et al., 2012). Single-stranded oligonucleotides (ssODNs) could be used as templates for ZFN-mediated precise gene editing, such as targeted point mutation, targeted genomic deletion and targeted insertion of small genetic elements (Chen et al., 2011). TALENs combined with ssODNs may be used for mimicking natural mutation of MSTN to produce novel “double muscle” livestock.
In this study, we describe methods of using simple ssODNs oligonucleotides in tandem with TALENs for editing MSTN gene in sheep.
MATERIALS AND METHODS
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless otherwise stated.
TALEN expression vectors and ssODNs oligonucleotides
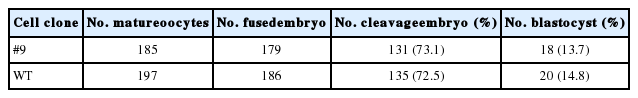
According to the sequence of sheep MSTN, a pair of TALENs were designed and assembled by the Golden Gate assembly method (Cermak et al., 2011). The TALENs were designed to target the sequence of exon 2 in the sheep MSTN gene. Each of the TALENs had a DNA binding domain recognizing 18 bases (Figure 1a). The ssODNs oligonucleotides were designed as described previously (Chen et al., 2011) and a stop codon (TAG) was inserted in ssODNs for disrupting MSTN expression (Supplementary Figure 1).
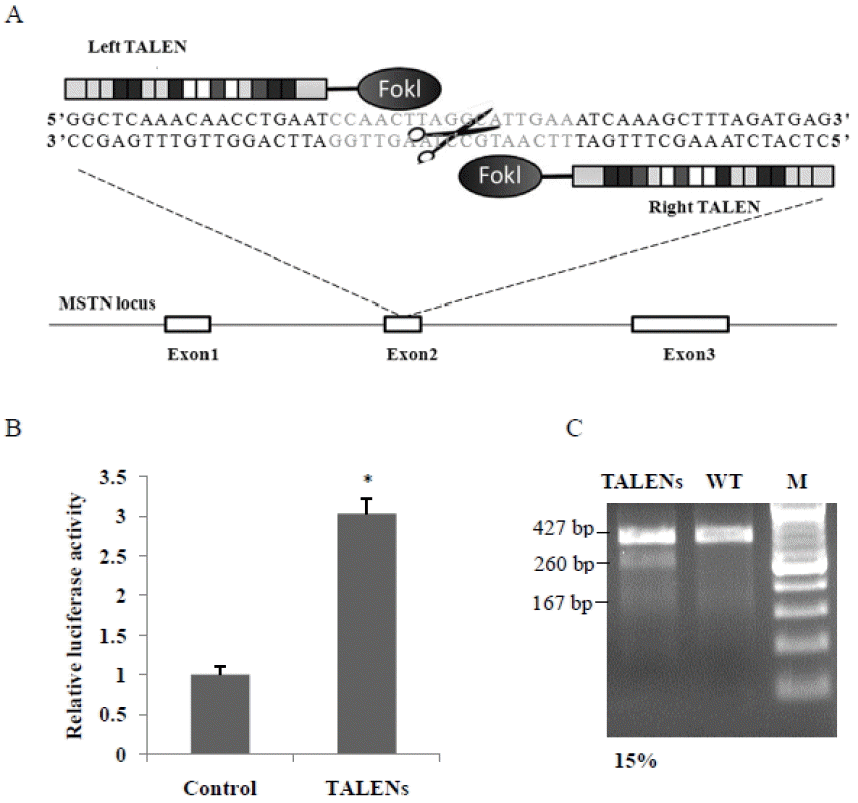
Design and activity of transcription activator-like effector nucleases (TALENs). (A) Schematic representation of TALENs targeting sheep myostatin (MSTN). The coding regions are indicated by white boxes. A TALEN consists of a nuclease domain (Fok I) and a DNA binding domain (TALE proteins), and the recognition sequences of the TALE proteins are blue. (B) TALENs activity was measured by luciferase SSA reporter assay. * p<0.01. (C) Surveyor nuclease assay for detecting TALEN activity. MSTN, myostatin; TALEN, transcription activator-like effector nuclease, polymerase chain reaction (PCR) products from TALEN-treated cells; SSA, single-strand annealing; WT, wild-type PCR products.
Luciferase single-strand annealing reporter assay
The luciferase single-strand annealing (SSA) reporter assay was performed as described previously (Wang et al., 2012). The luciferase SSA reporter construct contains two truncated luciferase fragments separated by a stop codon and MSTN TALEN target site. The 293T cells (American Type Culture Collection, Vanassas, MD, USA) were co-transfected with TALEN expression plasmid, SSA reporter plasmid (30 ng) (Viewsolid biotech, Beijing, China) and Renilla plasmid (5 ng). After 1 to 2 days of transfection, the relative luciferase activity was measured using a dual-luciferase assay system (Promega, Madison, WI, USA).
Surveyor nuclease assay
Sheep fibroblasts were isolated and cultured as previously described (Hu et al., 2013). Sheep fibroblasts were cultured in fresh Dulbecco’s Modified Eagle’s Medium with 10% fetal bovine serum (FBS) without antibiotics to achieve 80% to 90% confluency on the day of transfection. Sheep fibroblasts were transfected with 2 μg of each MSTN TALEN plasmid. At 72 hour post transfection, genomic DNA was extracted using the QuickExtract DNA extraction kit (Epicentre, Madison, USA) following the manufacturer’s protocol. Genomic DNA (700 ng) were used for polymerase chain reaction (PCR) using specific primers against MSTN (MSTN-F: 5′-GTT GCT TCT TTA AAT TTA GCT-3; MSTN-R: 5′-GAG ATT CTG TGG AGT GCT CAT-3′). PCR products were used for surveyor nuclease assay as described previously (Bedell et al., 2012). Briefly, PCR products were subjected to a re-annealing process to enable heteroduplex formation: 95°C for 10min, 95°C to 85°C ramping at −2°C/s, 85°C to 25°C at −0.25°C/s, and 25°C hold for 1 minute. After reannealing, products were treated with SURVEYOR nuclease and SURVEYOR enhancer S (Transgenomics, Omaha, NE, USA) following the manufacturer’s recommended protocol. The cleavage products were visualized by agarose gel (2%). The frequency of targeted gene mutation were calculated as previously described (Guschin et al., 2010).
Isolation of single cells clones
Sheep fetal fibroblasts were transfected with 2 μg of each MSTN TALEN plasmid or combined with 1 μg of ssODNs oligonucleotides using Nucleofector (Amaxa, Gaithersburg, MD, USA) according to the manufacturer’s protocol. The cells were incubated at 37°C (1 day) followed by 2 days at 30°C. Limited dilution was used in forming cell colonies and the cell concentration was about one cell/well in 96-well plates. Single cells were selected about 10 to 15 day after dilution culture and expanded cultured into 24-well culture dishes (Ni et al., 2014). When cells were nearly confluent, the cells clones were collected and divided into two parts. One part of the cells was used for mutation analysis by DNA sequencing. The other cells continued to be cultured.
DNA sequencing
Genomic DNA was isolated from TALEN-treated cell colonies. 700 ng of genomic DNA were used for PCR using specific primers against MSTN-F and MSTN-R as above. PCR products were gel purified and subjected to DNA sequence. DNA mutations were identified by sequence alignment between sequenced allele and wild type allele (Ni et al., 2014).
Somatic cell nuclear transfer
Sheep ovaries collected from a local slaughter house were transported to the laboratory in normal saline maintained between 27°C and 35°C. Cumulus-oocyte complexes were sucked out from follicles, maturated for 20 to 24 h in maturation medium (TCM-199 containing 20% FBS, 5 mg/mL of follicle stimulating hormone, 5 mg/mL of luteinizing hormone, and 1 mg/mL of estradiol). Mature oocytes were denucleated by aspirating the first polar body and fused with the donor cells enriched in G0 of the cell cycle, as the parameter of two DC pulses of 2.5 kv/cm for 10 ms each at 1 s apart, delivered by a BTX2001 Electro Cell Manipulator (BTX, San Diego, CA, USA). The reconstructed embryos were activated in medium (0.3 M mannitol, 0.1 mM MgCl2, and 0.05 mM CaCl2) supplemented with 10 mg/mL cycloheximede and 2.5 mg/mL cytochalasin D and cultured to form blastocysts at day 7 (Hu et al., 2013). Following activation, reconstructed embryos were transferred and cultured in SOFaa. A total of 70 embryos at the 2- to 4-cell stages were surgically transferred into 5 synchronized recipient ewes (10 to 15 embryos per recipient). Pregnancies were monitored by ultrasound scanning using a trans-abdominal linear probe at day 45.
RESULTS AND DISCUSSION
We designed and synthesized a pair of TALENs that targeted exon 2 of sheep MSTN (Figure 1a). Luciferase SSA reporter assay showed that co-transfection of MSTN TALEN and SSA reporter vector resulted in a 3-fold increase of luciferase activity (p<0.01) (Figure 1b), which suggested that the MSTN TALEN effectively induced DNA cleavage in HEK 293T cells. We further used surveyor nuclease test TALENs-mediated cleavage in sheep fibroblasts. Surveyor nuclease assay showed that the MSTN TALEN had high gene cleavage activity (15%) in sheep cells (Figure 1c).
Isolation of single cell-derived clones is required for developing methods of TALEN-mediated gene editing by SCNT. Sheep primary fibroblasts were transfected with TALENs only or TALENs and ssODN in tandem (TALEN/ssODNs). Colonies were isolated from the transfected fibroblasts by limited dilution. For TALEN/ssODNs group, we sequenced 35 colonies, and found that 4 colonies (11.4%) had the desired insertion of stop codon site in the MSTN gene (Figure 2a). For TALENs only group, 5 of 41 colonies (12.1%) carried some small deletions or insertion in the MSTN gene (Figure 2b). All mutated cells from TALENs only or TALEN/ssODNs groups were mono-allelic mutation. The efficiencies of TALEN-mediated gene editing with or without ssODNs were similar, suggested that ssODNs have no negative effect on TALENs activity. TALENs in tandem with ssODNs can be used for introducing precise gene editing in sheep primary fibroblasts.
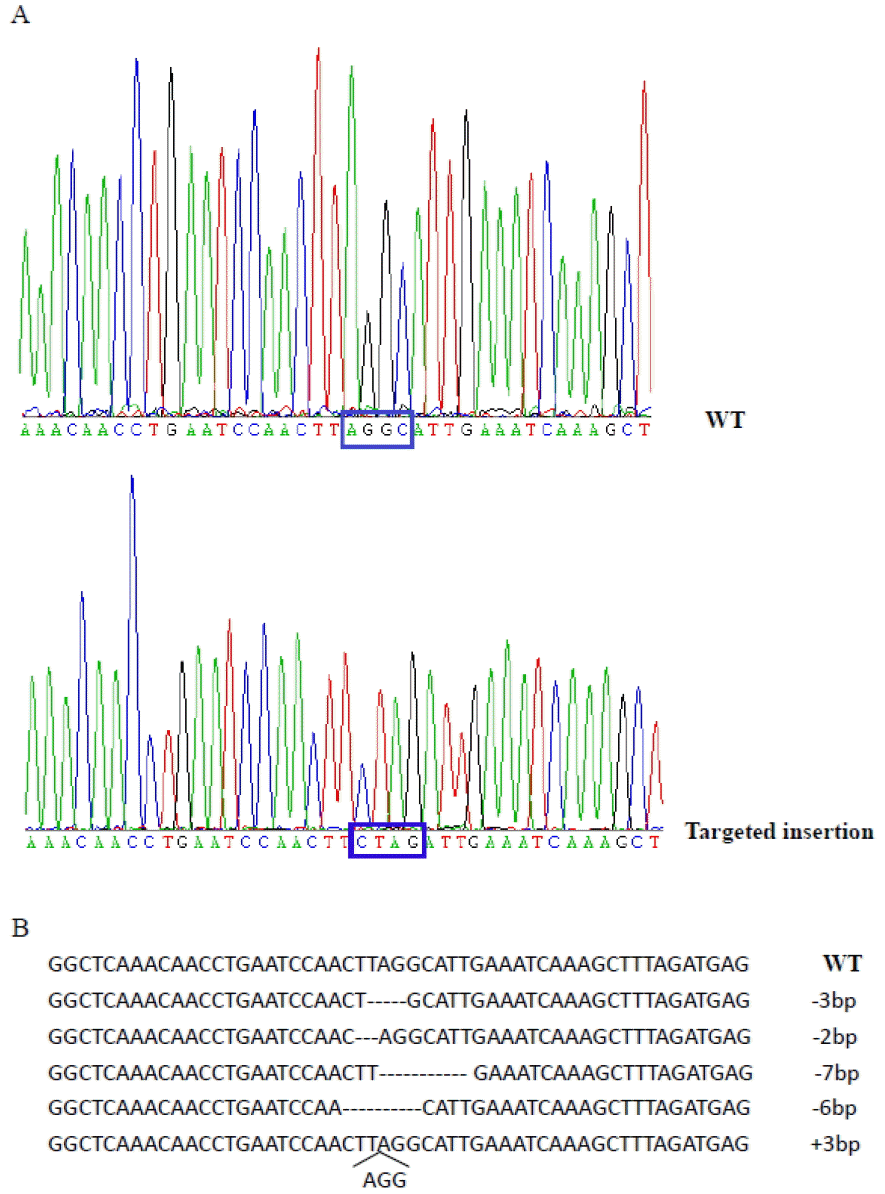
Sequence analysis of MSTN-modified cells. (A) MSTN sequence of cell colonies derived from TALEN/ssODNs-treated cells. WT, wild-type. Targeted insertion in #9 colonies: a stop codon (TAG) was targeted insertion into MSTN locus. (B) MSTN sequence of cell colonies derived from TALEN-treated cells. MSTN, myostatin; TALEN, transcription activator-like effector nuclease.
TALEN-mediated MSTN editing cells (#9 colonies) were used as nuclear donors for somatic cell cloning. The cleavage rates for TALEN-modified embryos were 73.1% (Table 1). The cloned embryos from TALEN-modified cells showed similar development rates to that from wild-type cells and that reported in other studies (Schnieke et al., 1997). One lamb was born at full term, but died soon after birth. The death was caused by dystocia at parturition. Postmortem revealed no histopathological abnormalities. DNA sequencing showed that the cloned lamb carried the desired insertion of stop codon site in the MSTN gene, which was identical with donor cell (#9 colonies) (Figure 2a).
Recently, Proudfoot et al. (2015) reported generation of MSTN gene-edited cattle and sheep by zygote injection of TALEN mRNA. Although direct modification of zygotic genomes may have some advantages, the strategy can result in mosaic or hypomorphic mutation of injected animals (Davies et al., 2013; Jao et al., 2013; Li et al., 2013), for which mutation may fail to transmit to offspring. In contrast, somatic cell modification followed by SCNT allow the isolation of mutant cells before the expense of animal production and ensure producing animals with expected gene modifications. We designed a pair of TALENs and one ssODNs oligonucleotides targeting sheep MSTN gene. Co-transfection of TALENs and ssODNs into primary fibroblasts enabled efficient isolation of MSTN-modified colonies. Normal embryo development from TALEN-modified cells suggested that TALEN-mediated sheep cells could be used for SCNT.
Our results show that TALENs/ssDNA achieve efficient targeted gene addition without antibiotic selection in sheep primary fibroblasts. Targeted point mutation or insertion of small genetic elements are very useful for producing gene-edited livestock. TALENs in tandem with sssODNs would be a potential tool for introducing desired mutations in the donor cells for animal cloning.
Supplementary materials
Sequence of the single-stranded DNA oligonucleotides (ssODNs) used to modify the MSTN locus. The ssODN is 60-mer and incorporate a stop codon (TAG) site for disrupting MSTN expression. The blue base represents the TALENs binding site. ssODN, single-stranded DNA oligonucleotide; MSTN, myostatin.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (NSFC) under Grant (31260534 and 31201800); the Bingtuan Five-year Special Projects under Grant (2012BB051), Outstanding youth (2014ZRKXJQ02) and Startup Foundation for Advanced Talents of Shihezi University under Grant (RCZX201318 and RCZX201428).
Notes
CONFLICT OF INTEREST
We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.