 |
 |
|
|
Abstract
Winter wheat is a suitable crop to be ensiled for animal feed and China has the largest planting area of this crop in the world. During the ensiling process, lactic acid bacteria (LAB) play the most important role in the fermentation. We investigated the natural population of LAB in whole-crop wheat (WCW) and examined the quality of whole-crop wheat silage (WCWS) with and without LAB inoculants. Two Lactobacillus plantarum subsp. plantarum strains, Zhengzhou University 1 (ZZU 1) selected from corn and forage and grass 1 (FG 1) from a commercial inoculant, were used as additives. The silages inoculated with LAB strains (ZZU 1 and FG 1) were better preserved than the control, with lower pH values (3.5 and 3.6, respectively) (p<0.05) and higher contents of lactic acid (37.5 and 34.0 g/kg of fresh matter (FM), respectively) (p<0.05) than the control. Sixty LAB strains were isolated from fresh material and WCWS without any LAB inoculation. These LAB strains were divided into the following four genera and six species based on their phenotypic, biochemical and phylogenetic characteristics: Leuconostoc pseudomesenteroides, Leuconostoc citreum, Weissella cibaria, Lactococcus lactis subsp. lactis, Lactobacillus buchneri, and Lactobacillus plantarum subsp. plantarum. However, the prevalent LAB, which was predominantly heterofermentative (66.7%), consisted of Leuconostoc pseudomesenteroides, Leuconostoc citreum, Weissella cibaria, and Lactobacillus buchneri. This study revealed that most of isolated LAB strains from control WCWS were heterofermentative and could not grow well at low pH condition; the selective inoculants of Lactobacillus strains, especially ZZU 1, could improve WCWS quality significantly.
Winter wheat (Triticum aestivum L.) is one of the most popular crops in the world, including China, which is the largest producer that produced almost 111 million tons of this crop every year in the world. The North Plain, especially the Henan Province, producing about two-thirds of the total wheat production, is the most important winter wheat growing area in China (Lu et al., 2013). As the crop can be grown in a wide range of climates and soil conditions, and has relative high contents of dry matter (DM) and water soluble carbohydrate (WSC) which could improve ensiling process. Some countries, such as northern Europe, began to explore the feeding value as the whole-crop wheat silage (WCWS) (Ashbell et al., 1985; Adamson et al., 1992; Ashbell et al., 1997). The study of Wang et al. (2013) found that the alternative of corn silage to WCWS had a tendency to raise milk production of cows. The crude protein content of silage prepared with whole-crop wheat (WCW) in milk-ripe stage was 1% to 2% higher than that of whole corn silage, while neutral detergent fiber (NDF) and acid detergent fiber (ADF) were close. With the improvement of people’s living standard and the adjustment of economic structure, some areas in North China are transforming traditional farming pattern to the cultivating crops for livestock; besides, some wheat with relatively poor quality is not suitable for making food. Therefore, it is necessary for us to investigate the potential ability to develop the WCWS in China.
Ensiling is the most commonly preservation method for forage crop and is fermented by a complex consortium of microorganisms, predominantly LAB, which convert WSC into sufficient organic acids, mainly lactic acid, under anaerobic condition. As a result, pH decreases and undesirable microorganisms are inhibited (Cai, 1999; Pang et al., 2012). Many LAB inoculants and chemical additives have been developed, because LAB inoculants are safe and easy to apply, non-corrosive to machine, do not pollute the environment and are regarded as natural products (Filya et al., 2000). Thus, many LAB-containing additives, which include Lactobacillus (L.) plantarum, L. rhamnosus, L. acidophilus, Pediococcus acidilactici, and Enteroccus faecium have been developed and are available (Cai et al., 1998). For example, Xie et al. (2012) found inoculating Lactobacillus parafarraqinis could enhance the aerobic stability of wheat silages at different maturity. These inoculants may involve accelerating the process of silage fermentation and dominating lactic acid fermentation.
To our knowledge, from microbiological perspective, little information is available on the microbial ecology of fresh wheat material, especially the inhabited LAB, which determines whether inoculant needs to be used. Therefore, the present study was to investigate the microorganism constitution of fresh wheat material and control WCWS, identify isolates with physiological and biochemical test, and determine selected representative strains at the molecular level using 16S rRNA and recA sequence analysis
The whole crop wheat (Yumai 19) was collected at the milk stage from a farmland in Henan Province, China in the May, 2012. Two L. plantarum subsp. plantarum strains Zhengzhou University 1 (ZZU 1) isolated from corn and forage and grass 1 (FG 1) from a commercial inoculant (Chikuso-1) were used, and they were incubated in lactobacilli de Man, Rogosa, Sharpe (MRS) broth overnight. After incubation, the optical density of the suspension at 700 nm was adjusted to 0.40 using sterile 0.85% NaCl solution just before ensiling. Then 100 μL diluted LAB solution per 100 gram of fresh matter (FM) was applied. Silages were prepared using a small scale system, approximately 100 g portions of forge material chopped into about 20-mm length and packed into plastic bags (N-9, Asahi Kasei Co., Ltd., Tokyo, Japan). Experimental treatments included: control silage without LAB, the WCW+FG 1 silage and WCW+ZZU 1 silage. The bag silos were vacuumized and sealed with Sharp Vacuum Seal/Package (SQ-202, Sharp Co., Ltd., Tokyo, Japan). Eight bags per treatment were made. The plastic bags were stored at room temperature (28.0°C to 35.0°C) and three bags on day 30 of each treatment were opened for evaluations of silage fermentation.
The samples (10 g) were blended with 90 mL of sterilized water, and serially diluted from 10−1 to 10−5 in sterilized water. The number of LAB was measured by plate count on MRS agar incubated at 30°C for 48 h under anaerobic conditions (DG 250/min MACS; Don Whitley Science, Shipley, England). Coliform bacteria were counted on blue light broth agar (Nissui-Seiyaku Co., Ltd., Tokyo, Japan), incubated at 30°C for 48 h. Molds and yeasts were counted on potato dextrose agar (Nissui-Seiyaku Ltd., Japan), incubated at 30°C for 24 h, and yeasts were distinguished from molds and other bacteria by colony appearance and the observation of cell morphology. Bacilli and aerobic bacteria were counted on nutrient agar (Nissui Ltd., Japan), incubated at 30°C for 24 h under aerobic conditions. Colonies were counted as viable numbers of microorganisms in CFU/g of FM. Three repetitions of each sample were analyzed during the isolation process, and almost all of these from same sample were similar, therefore, one sample data were used in this experiment. Each LAB colony was isolated and purified twice by streaking on MRS agar plates. Pure cultures were grown on MRS agar at 30°C for 24 h, and then the purified strains were stored at −80°C in nutrient broth (Difco) for further examination.
Morphological, physiological and biochemical tests of LAB morphology and Gram-staining response were examined after 24 h of incubation on MRS agar. Catalase activity and gas production from glucose were determined using the methods of Kozaki et al. (1992). The LAB strains producing gas from glucose were regarded as heterofermentative type while the LAB strains producing no gas were regarded as homofermentative type. Growth at different temperatures was observed in MRS broth after incubation at 5°C, 10°C, and 15°C for 14 days, and at 45°C and 50°C for 7 days. Growth at pH 3.0, 3.5, 4.0, 4.5, 5.0, 6.0, and 8.0 was observed in MRS broth after incubation at 30°C for 7 days. Salt tolerance of LAB was tested in MRS broth containing 3.0% and 6.5% NaCl at 30°C for 2 days. Carbohydrate assimilation and fermentation of 49 compounds with one control were identified on Analytic Products INC (API) 50 CH strips (bioMerieux, Tokyo, Japan).
Cells grown at 30°C for 24 h in MRS agar were used for 16S rRNA gene sequence. The 16S rRNA gene was amplified by polymerase chain reaction (PCR) in a thermal cycler. The sequences of the PCR products were determined directly with a sequencing kit using the prokaryotic 16S ribosomal DNA universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) (Pang et al., 2011a). Sequence similarity searches were performed using the DNA Database of Japan (DDBJ) and the basic local alignment search tool. The sequence information was then imported into the Sequencher which contains Clustal X 1.81 software program (Hitachi Software Engineering Co., Tokyo, Japan) for assembly and alignment. The 16S rDNA sequences of ZZU strains were compared with sequences from type LAB strains held in the DDBJ. Nucleotide substitution rates (Kunc values) were calculated and phylogenetic trees were constructed using the neighbor-joining method (Kimura et al., 1972). Bacillus subtilis NCDO 1769T was used as an outgroup organism. The topologies of trees were evaluated using bootstrap analysis of the sequence data with molecular evolutionary genetic analysis (MEGA) 4 software, based on 1,000 random resampling (Tamura et al., 2007). These sequences were aligned with the type published sequences from DDBJ, GenBank and the European Molecular Biology Laboratory (EMBL).
For further discrimination of strains in the L. plantarum group, a multiplex PCR assay was performed with the recA gene-based primers: paraF (5′-GTCACAGGCATTACGAA AAC-3′), pentF (5′-CAGTGGCGCGGTTGATATC-3′), planF (5′-CCGTTTATGCGGAACACCTA-3′), and pREV (5′-TCGGGATTACCAAACATCAC-3′). The PCR mixture and amplifications were performed as described by Torriani et al. (2001).
The nucleotide sequences for the 16S rDNA described in this report were deposited with GenBank under accession nos. AB30322, AB30323, AB30324, AB30325, AB30326, AB30327 and AB30328 for the strains ZZU 16, ZZU 20, ZZU 23, ZZU 3, ZZU 8, ZZU 12, and ZZU 21, respectively.
At the 30 day of ensiling, three bags of WCWS per treatment were opened for measuring fermentation quality and chemical composition. The pH was measured with a glass electrode pH meter (pH 213; HANNA, Padova, Italy). The ammonia-N was determined by steam distillation of the filtrates. The organic acid contents were measured by HPLC (1200series; Agilent, Santa Clara, CA, USA) according to the method described by Cai et al. (1999). The WCWS samples were dried in a forced-air oven at 65°C for 48 h and ground to pass a 1-mm screen with a Wiley mill (ZM200, Retsch GmbH, Haan, Germany). Dry matter, ether extract and organic matter were analyzed according to AOAC Methods 934.01, 920.39, and 942.05, respectively (AOAC, 1990). The NDF and ADF were analyzed by the method of Van Soest et al. (1991). Wet silage (10 g) was homogenized with 90 mL sterilized distilled water.
Data on the fermentation quality and chemical composition of the 30 d silages were analyzed by analysis of variance, and the significance of differences among means was tested by the general linear model procedures of SAS (SAS Institute Inc., Cary, NC, USA). The statistical model included the main factor of treatment (3 levels) and the random factor of silage within treatment. Statistical analysis was performed using one-way analysis of variance with post hoc Duncan test; p<0.05 was considered significant.
As shown in Table 1 and 2, all of the LAB strains isolated from the control WCWS were Gram-positive and catalase-negative and could not grow at temperatures below 5°C or above 50°C. No LAB strains could grow both at pH 3.0 and in 6.5% NaCl, except strain ZZU 20. According to the morphological, physiological and biochemical properties, the isolated LAB strains were divided into six groups (A-F). The strains in group A (ZZU 3 and 21) produced acid from galactose, melibiose, trehalose, D-raffinose, and D-turanose and could not grow in 6.5% NaCl, while the strain in group C (ZZU 8) could not ferment those carbohydrates, but could grow in 6.5% NaCl. The group B strain ZZU 16 could not produce acid from ribose or D-xylose, making it different from groups A and C. Unlike the strains in groups A, B, and C, the group D strain (ZZU 12) did not produce gas from glucose and did not grow at pH 4.5. All of the strains in group E (ZZU 23) and group F were rod-shaped, but those in group E were homofermentative, while those in group F were heterofermentative.
Phylogenetic trees of 7 representative strains constructed from evolutionary distances using the neighbour-joining method are shown in Figure 1 and 2. Following the 16S rDNA sequence analysis, the strains in all groups were placed in clusters based on their genera, Leuconostoc, Lactococcus, Weissella, and Lactobacillus. Strains in groups A and B were placed in the cluster of Leuconostoc (Ln.). Ln. pseudomesenteroides JCM 9696T and Ln. citreum JCM 9698T were the most closely related species, indicated by a more than 99.7% similarity in their 16S rRNA gene sequences. Therefore, the strains in groups A and B belonged to Ln. pseudomesenteroides and Ln. citreum, respectively. The representative strain in group C was clearly defined as Weissella (W.) cibaria supported with a 100% bootstrap value. The group D strain was found to be Lactococcus (Lc.) lactis because group D formed a very well-defined cluster with the following three subspecies: Lc. lactis subsp. hordniae, Lc. lactis subsp. lactis, and Lc. lactis subsp. cremoris; the group D strain was further ascribed to the subspecies lactis on the phylogenetic tree, with a 100% bootstrap value and 99.9% similarity for the 16S rRNA gene sequence supporting its monophyly. The strains of group E and F were all placed in the cluster of Lactobacillus; group E strains were grouped on the phylogenetic tree with L. plantarum subsp. plantarum, L. plantarum subsp. argentoratensis, L. pentosus and L. paraplantarum, and the representative strain of group F was identified as L. buchneri, with 99.8% similarity for the 16S rRNA gene sequence. In the fresh matter, only W. cibaria was found.
Amplification products obtained from the recA gene of the representative strains of group E and the type strain are shown in Figure 3. The group E strains and L. plantarum subsp. plantarum JCM 1149T produced 318 bp products. Thus, all of the group E strains were clearly identified as L. plantarum subsp. plantarum.
The microbiological composition of silages after 30 days of fermentation are shown in Table 3. The numbers of coliform bacteria in the treated silages decreased to a low level and were too few (<103 colony-forming unit [CFU]/g) to count after 30 days of fermentation. The counts of aerobic bacteria and yeasts decreased to a lower level (104 CFU/g) than that in the control. Table 4 shows the fermentation quality and chemical composition of the silages after 30 days of fermentation. Propionic acid and n-Butyric acid were below the detectable level (0.01 g/kg of FM) in all of the silages. Compared with the control, the silages inoculated with LAB (ZZU 1 and FG 1) were preserved well with relatively lower pH values (3.5 and 3.6, respectively) and higher lactic acid contents (37.5 and 34.0 g/kg of FM, respectively) (p<0.05). In addition, the DM for the ZZU 1-treated silage was higher (p<0.05) than that for the control, and the DM for the FG 1-treated silage was not significantly higher than that for the control (p<0.05).
Wheat is cultivated beyond 240 million hectares, larger than any other crops in the developing country. As one of the top producers of world, China’s share is almost one-sixth of the world production of wheat (Weinberg et al., 2010). Wheat is becoming more and more popular to be used as silage in the world, and there are some advantageous characteristics of wheat as a forage crop: it can adapt different kinds of soils and climate conditions; the moisture is adequate for ensiling when it is harvested in the spring; if wheat is ensiled properly with other crop grains, it can increase milk production. In addition, with the increasing price of corn caused by biofuel production, wheat silage could be an alternative to corn silage (Weinberg et al., 2009). The fermentation process of LAB for several forage crops and grasses have been described (Cai, 1999). This study provides a unique opportunity to investigate microbial flora of wheat silage, and study the effects of fermentation quality and chemical composition on the wheat silage by LAB inoculant.
In order to reproduce the fermentation process and exactly analyze fermentation quality, a small-scale fermentation method was developed (Tanaka et al., 1995). This method employs about 100 to 1,000 g of portions of forage material to pack into plastic bag silo, then the bag silo was sealed with a vaccum sealer. The plastic silo was made by using the laminated plastic film of nylon and polyethylene and silage prepared with this method can store well more than one year (Cao et al., 2011; Chen et al., 2013). This method can reproduce the silage fermentation process, and greatly contribute to the study of silage fermentation principle. In addition, in some comparative tests of silage fermentation using various silo, the results of silage fermentation by using small-scale fermentation method were same or similar (not significantly) to other silo methods such as bottle, jar, role bale, and bunk silo (Tanaka et al., 1995; Müller, 2005).
In this study, all the isolated LAB were characterized by Leuconostocs; W., Weissella; L., Lactococcus; E., Enterococcus. the physical procedure, API 50 CH strips, and for the purpose of identifying LAB isolates at the species level, molecular phylogeny analysis was employed and phylogenetic trees were constructed based on the 16S rDNA sequences from evolutionary distances by the neighbor-joining method, which led to delineation of 6 groups of isolates, and strains in group A, B, and C were all identified to clear phylogenetic position (Eitan et al., 2006). Group D representative strain ZZU 12 was clearly ascribed to Lc. lactis cluster containing three subspecies: Lc. lactis subsp. cremoris, Lc. lactis subsp. lactis, and Lc. lactis subsp. hordniae (Figure 1). However, 16S rDNA sequence analysis could not differentiate strains at the subspecies level or efficiently classify interspecies relationships (Stackebrandt et al., 1994), combine with carbohydrate fermentation patterns nearly identical to that type strain of Lc. lactis subsp. lactis 5805T excepting maltose and N-acetyl glucosamine, but their ribose, D-Xylose, mannitol and amygdaline fermentation patterns differed from those of the other two subspecies (Table 2). In addition, the strain ZZU 12 could grow in 0.1% bile salt, which made it different from the other two subspecies (Kim et al., 1999), and therefore, group D strains could be identified as Lc. lactis subsp. lactis. The same difficulty exists in the group E, members L. plantarum cluster including L. pentosus, L. plantarum subsp. plantarum, L. plantarum subsp. argentoratensis, and L. paraplantarum, which have very similar 16S rDNA gene sequences that differ only by 2 bp. The PCR product of recA gene is regarded as an efficient way for differentiating the distantly related species (Torriani et al., 2001). The PCR amplification analysis of partial recA gene products permitted a clear distinction between the four type strains and group E strains. The strain ZZU 23, represented strain of group E, had the same product (318 bp) as the L. plantarum subsp. plantarum. Therefore, strain ZZU 23 could be identified as L. plantarum subsp. plantarum.
Different species and the characteristics of epiphytic LAB might change and influence fermentation process and silage quality. During early fermentation phase, WSC are converted into CO2, heat and water by both plant cells and aerobic microbes, lactic acid-producing cocci, e.g. lactococci, leuconostocs, weissella, streptococci, pediococci, and enterococci, grow vigorously and start lactate fermentation by creating an anaerobic environment suitable for the development of lactobacilli (Ennahar et al., 2003). In the present study, the various groups presumably represented 4 genera including one rod (Lactobacillus) and three cocci (Lactococcus, Weisslla, and Leuconostoc). In contrast with cocci that could not grow at low pH values, lactobacilli play a more important role in promoting lactic acid fermentation for a longer time, leading to a sharp drop in pH. Many studies have indicated that inoculating forage with homofermentative lactobacilli such as L. casei or L. plantarum has beneficial effects on increasing the amount of lactic acid content and improving silage quality. Heterofermentative LAB can improve silage fermentation quality to the some extent in some cases, but their efficiency is not as good as homofermentative ones (Cai, 1999; Pang et al., 2011b).
It was reported that epiphytic lactobacilli must reach a level of at least 105 CFU/g FM to ensure successful silage storage (Ennahar et al., 2003; Pang et al., 2012), while in the present study, the number of LAB were below 104 CFU/g of FM, and the counts of aerobic bacteria, coliform bacteria and yeasts were higher than 104 CFU/g of FM in the fresh wheat material (data not shown). When LAB fail to produce adequate lactic acid in the process of silage fermentation to reduce pH and suppress the growth of harmful bacteria, the resulting silage will be of poor quality. In the present study, the control silage show higher numbers of coliform bacteria after day 5 of fermentation. The most plausible explanation lies in the fermentation quality. The control silages did not improve silage quality, if the silage pH does not decline to <4.0, the growth of coliform bacteria and aerobic bacteria is not inhibited (Cai et al., 1998; 2003).
The objectives of inoculating silage with LAB are to accelerate the production of lactic acid and lower the pH values, and inhibit the reproduction of harmful bacteria (Ashbell et al., 1997). Many commercial LAB inoculants are selected to add the ensilaged forage to predominate and outnumber the naturally epiphytic LAB present in the forage. They may be dependent on several characteristics, such as their capacity to dominate the natural microflora in the forages and to produce lactic acid rapidly due to effective fermentation (Hellings et al., 1985). This research found that all the harmful microorganisms and pH values of the treated silages declined rapidly than the control; the values of pH and ammonia-N content of ZZU 1 treated silages were relatively equal with that treated with FG 1. FG 1 is a popular commercial inoculant both in China and Japan. Compared with the FG 1, strain ZZU 1 is better than commercial strain FG 1 in some cases, such as it can grow at the sanity of 6.5% and pH 3.0. That may mean ZZU 1 strain has potential value being as a commercial inoculant for forage crop ensiling, and more experiments about its effectiveness in other crops are underway.
The isolated LAB strains from the fresh wheat and control WCWS were mostly heterofermentative and did not have a strong surviving ability in low pH condition. The treated silages with L. plantarum subsp. plantarum strains ZZU 1 and FG 1 were preserved well, which shows the additive of LAB, especially the lactobacilli, is necessary for wheat silage. In addition, the strain ZZU 1 showed a potential ability for developing as an excellent additive.
ACKNOWLEDGMENTS
This work was supported by Foundation of Henan Science and Technology Committee (Grant No. 132300410054).
Figure 1
Phylogenetic tree showing the relative positions of Leuconostoc, Weissella, Lactococcus, and Enterococcus species, as inferred by the neighbor-joining method with 16S rRNA gene sequences. B. subtilis is used as an outgroup. Bootstrap values for a total of 1,000 replicates are shown at the nodes of the tree. The bar indicates 1% sequence divergence. Knuc, nucleotide substitution rates. Ln.,
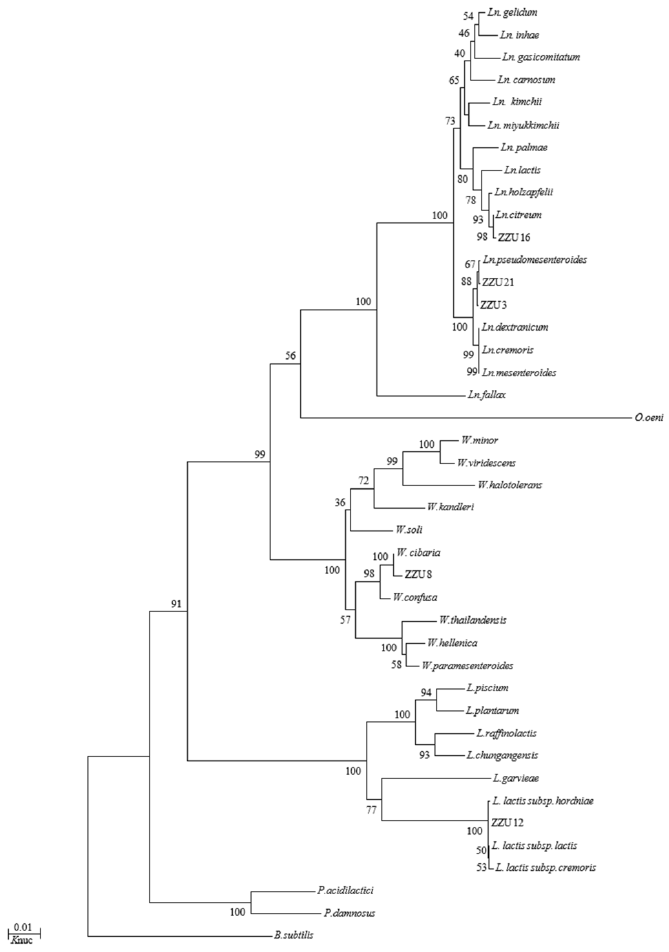
Figure 2
Phylogenetic tree showing the relative position of L. plantarum, L. brevis, and L. paraplantrum species as inferred by the neighbor-joining method with 16S rRNA gene sequences. B. subtilis is used as an outgroup. Bootstrap values for a total of 1,000 replicates are shown at the nodes of the tree. The bar indicates 1% sequence divergence. Knuc, nucleotide substitution rates.
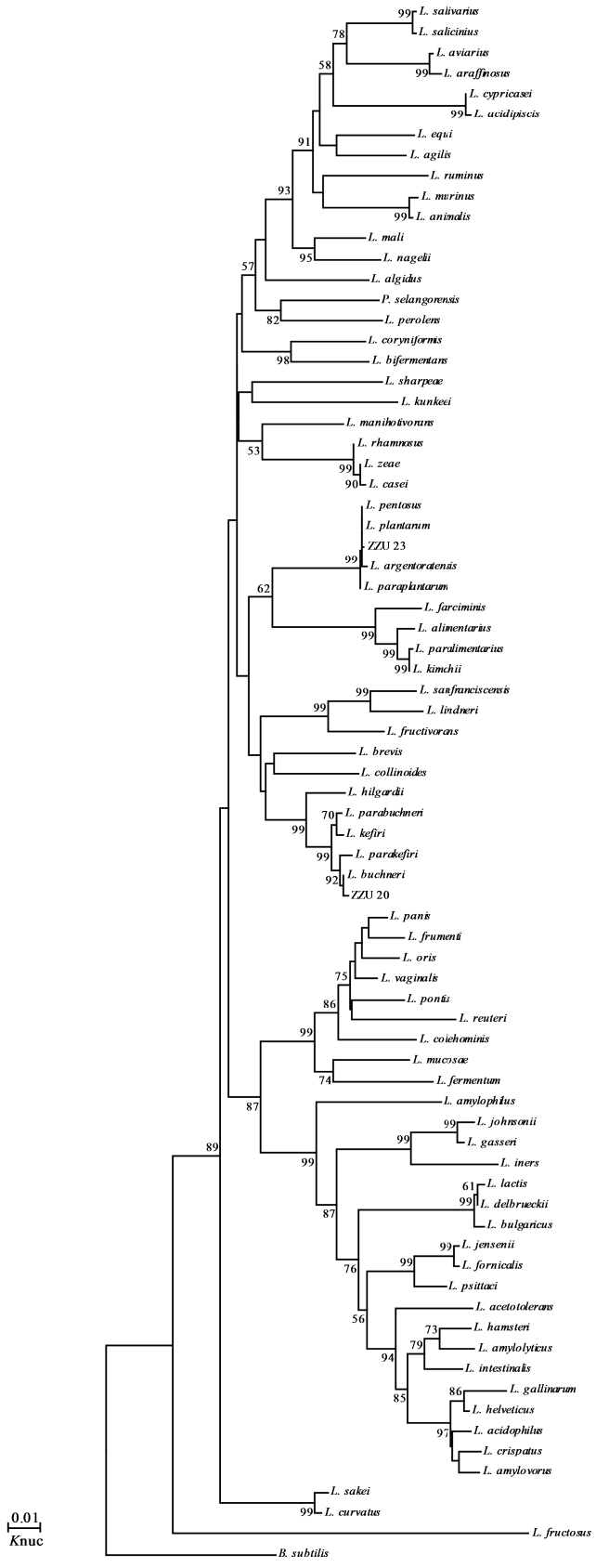
Figure 3
Amplification products obtained from the recA multiplex assay. Lane M contained a 600 bp PLUS DNA ladder (Tiangen Biotech Co, Ltd., Beijing, China). Lanes 1, 2, 3, 4, and 5, PCR amplication products from L. casei JCM 16167T (negative control), L. paraplantarum JCM 12533T, L. pentosus JCM 1558T, L. plantarum subsp. plantarum JCM 1149T, and L. plantarum subsp. argentoratensis JCM 16169T, respectively; Lane 6, PCR amplification product from ZZU 23. PCR, polymerase chain reaction.
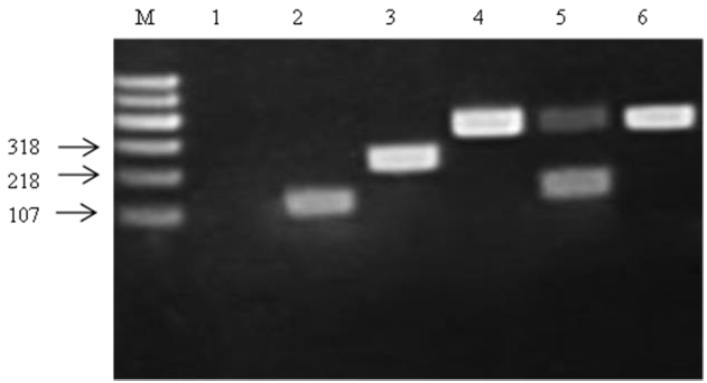
Table 1
Characteristics of isolated LAB strains from the control WCWS
Table 2
API 50 CH fermentation patterns of isolated LAB from the control WCWS
API, Analytic Products INC; LAB, lactic acid bacteria; WCWS, whole-crop wheat silage; JCM, Japan Collection of Microoganisms; ZZU, Zhengzhou University; T, Type strains.
Table 3
Microorganisms of whole-crop wheat ensiled for 30 d
| Counts (log CFU/g of FM) of viable microorganisms | ||||
|---|---|---|---|---|
|
|
||||
| Coliform bacteria | Aerobic bacteria | Yeasts | LAB | |
| Control | 4.1 | 6.2a | 5.6a | 4.7b |
| ZZU 1 | ND | 4.9b | 4.5b | 5.7a |
| FG 1 | ND | 4.1b | 4.9b | 4.1b |
| SEM | - | 0.5 | 0.6 | 0.3 |
Table 4
Fermentation quality and chemical composition of whole-crop wheat silages ensiled for 30 d1
REFERENCES
Adamson AH, Reeve A. 1992. Nutritional evaluation of whole-crop wheat. Whole-Crop Cereals. Stark BA, Wilkinson JM, editorsChalcombe Publications; Aberystwyth, UK: p. 85–96.
AOAC. 1990. Official Methods of Analysis. 15th edAssociation of Official Analytical Chemists; Arlington, VA, USA:
Ashbell G, Theune HH, Sklan DJ. 1985. Ensiling whole wheat at various maturation stages: Changes in nutritive ingredients during maturation and ensiling and upon aerobic exposure. J Agric Food Chem 33:1–4.

Ashbell G, Weinberg ZG, Bruckental I, Tabori K, Sharet N. 1997. Wheat silage: Effect of cultivar and stage of maturity on yield and degradability in situ. J Agric Food Chem 45:709–712.

Cai Y. 1999. Identification and characterization of Enterococcus species isolated from forage crops and their influence on silage fermentation. J Dairy Sci 82:2466–2471.


Cai Y, Benno Y, Ogawa M, Ohamomo S, Kumai S, Nakase T. 1998. Influence of Lactobacillus spp. from an inoculant and of Weissellla and Leuconostoc spp. from forage crops on silage fermentation. Appl Environ Microbiol 64:2982–2987.



Cai Y, Fujita Y, Murai M, Yoshida N, Kitamura A, Miura T. 2003. Application of lactic acid bacteria (Lactobacillus plantarum Chikuso-1) for silage preparation of forage paddy rice. J Jpn Soc Grassland Sci 49:477–485.
Cao Y, Cai Y, Takahashi T, Yoshida N, Tohno M, Uegaki R, Nonaka K, Terada F. 2011. Effect of lactic acid bacteria inoculant and beet pulp addition on fermentation characteristics and in vitro ruminal digestion of vegetable residue silage. J Dairy Sci 94:3902–3912.


Chen MM, Liu QH, Xin GR, Zhang JG. 2013. Characteristics of lactic acid bacteria isolates and their inoculating effects on the silage fermentation at high temperature. Lett Appl Microbiol 56:71–78.


Ennahar S, Cai Y, Fujita Y. 2003. Phylogenetic diversity of lactic acid bacteria associated with paddy rice silage as determined by 16S ribosomal DNA analysis. Appl Environ Microbiol 69:444–451.



Eitan BD, Shapiro OH, Siboni N, Kushmaro A. 2006. Advantage of using inosine at the 3′ termini of 16S rRNA gene universal primers for the study of microbial diversity. Appl Environ Microbiol 72:6902–6906.



Filya I, Ashbell G, Hen Y, Weinberg ZG. 2000. The effect of bacterial inoculants on the fermentation and aerobic stability of whole crop wheat silage. Anim Feed Sci Technol 88:39–46.

Hellings P, Bertin G, Vanbelle . 1985. Effect of lactic acid bacteria on silage fermentation. In : Proceedings 15th International Grassland Congress; Kyotosyuppan, Kyoto, Japan. p. 932–933.
Kim WS, Ren J, Dunn NW. 1999. Differentiation of Lactococcus lactis subspecies lactis and subspecies cremoris strains by their adaptive response to stresses. FEMS Microbiol Lett 171:57–65.


Kimura M, Ohta T. 1972. On the stochastic model for estimation of mutational distance between homologous proteins. J Mol Evol 2:87–90.


Kozaki M, Uchimura T, Okada S. 1992. Experimental Manual of Lactic Acid Bacteria. Asakurashoten; Tokyo, Japan:
Lu C, Fan L. 2013. Winter wheat yield potentials and yield gaps in the North China Plain. Field Crops Res 143:98–105.

Müller CE. 2005. Fermentation patterns of small-bale silage and haylage produced as a feed for horses. Grass Forage Sci 60:109–118.

Pang H, Qin G, Tan Z, Li Z, Wang Y, Cai Y. 2011a. Natural populations of lactic acid bacteria associated with silage fermentation as determined by phenotype, 16S ribosomal RNA and recA gene analysis. Syst Appl Microbiol 34:235–241.


Pang H, Tan Z, Qin G, Wang Y, Li Z, Jin Q, Cai Y. 2012. Phenotypic and phylogenetic analysis of lactic acid bacteria isolated from forage crops and grasses in the Tibetan Plateau. J Microbiol 50:63–71.


Pang H, Zhang M, Qin G, Tan Z, Li Z, Wang Y, Cai Y. 2011b. Identification of lactic acid bacteria isolated from corn stovers. Anim Sci J 82:642–653.


Stackebrandt E, Goebel BM. 1994. A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Evol Microbiol 44:846–849.

Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA 4: Molecular Evolutionary Genetics Analusis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599.


Tanaka O, Ohmomo S. 1995. A simple method of laboratory silage fermentation by using a plastic pouch for packaging. Grassland Sci 41:55–59.
Torriani S, Felis GE, Dellaglio F. 2001. Differentiation of Lactobacillus plantarum, L. pentosus, and L. paraplantarum by recA Gene Sequence Analysis and Multiplex PCR Assay with recA Gene-Derived Primers. Appl Environ Microbiol 67:3450–3454.



Van Soest PJ, Robertson JB, Lewis BA. 1991. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J Dairy Sci 74:3583–3597.


Wang Q, Tang B, Han Z. 2013. The effects of the different silage on milk yield and composition in dairy cows. Dairy Cows 9:52–54.
Weinberg ZG, Khanal P, Yildiz C, Chen Y, Arieli A. 2010. Effects of stage of maturity at harvest, wilting and LAB inoculant on aerobic stability of wheat silages. Anim Feed Sci Technol 158:29–35.

- TOOLS
-
METRICS

- Related articles
-
Bacterial Inoculant Effects on Corn Silage Fermentation and Nutrient Composition2009 July;22(7)





 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print




