Aitman TJ, Dong R, Vyse TJ, Norsworthy PJ, Johnson MD, Smith J, Mangion J, Roberton-Lowe C, Marshall AJ, Petretto E, et al. 2006. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature 439:851ŌĆō855.


Aldred PMR, Hollox EJ, Armour JAL. 2005. Copy number polymorphism and expression level variation of the human ╬▒-defensin genes
DEFA1 and
DEFA3. Hum Mol Genet 14:2045ŌĆō2052.


Alterovitz G, Ramoni MF. 2010. Knowledge Based Bioinformatics: From Analysis to Interpretation. John Wiley & Sons, Ltd; Chichester, UK:
Alvarez CE, Akey JM. 2012. Copy number variation in the domestic dog. Mamm Genome 23:144ŌĆō163.


Bolstad B. preprocessCore: A collection of pre-processing functions. R package version 1.
Casta├▒o-Rodr├Łguez N, Diaz-Gallo LM, Pineda-Tamayo R, Rojas-Villarraga A, Anaya JM. 2008. Meta-analysis of
HLA-DRB1 and
HLA-DQB1 polymorphisms in Latin American patients with systemic lupus erythematosus. Autoimmun Rev 7:322ŌĆō330.


Cobb JP, Mindrinos MN, Miller-Graziano C, Calvano SE, Baker HV, Xiao W, Laudanski K, Brownstein BH, Elson CM, Hayden DL, et al. 2005. Application of genome-wide expression analysis to human health and disease. Proc Natl Acad Sci USA 102:4801ŌĆō4806.



Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. 2003. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4:R60


Feuk L, Carson AR, Scherer SW. 2006. Structural variation in the human genome. Nat Rev Genet 7:85ŌĆō97.


Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, Catano G, Nibbs RJ, Freedman BI, Quinones MP, Bamshad MJ, et al. 2005. The influence of
CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science 307:57141434ŌĆō1440.


Guryev V, Saar K, Adamovic T, Verheul M, van Heesch SAAC, Cook S, Pravenec M, Aitman T, Jacob H, Shull JD, Hubner N, Cuppen E. 2008. Distribution and functional impact of DNA copy number variation in the rat. Nat Genet 40:538ŌĆō545.


Henrichsen CN, Chaignat E, Reymond A. 2009a. Copy number variants, diseases and gene expression. Hum Mol Genet 18:R1ŌĆōR8.


Henrichsen CN, Vinckenbosch N, Z├Čllner S, Chaignat E, Pradervand S, Sch├╝tz F, Ruedi M, Kaessmann H, Reymond A. 2009b. Segmental copy number variation shapes tissue transcriptomes. Nat Genet 41:424ŌĆō429.


Inagaki T, Suzuki S, Miyamoto T, Takeda T, Yamashita K, Komatsu A, Yamauchi K, Hashizume K. 2003. The retinoic acid-responsive proline-rich protein is identified in promyeloleukemic HL-60 cells. J Biol Chem 278:51685ŌĆō51692.


Jaenisch R, Bird A. 2003. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat Genet 33:245ŌĆō254.


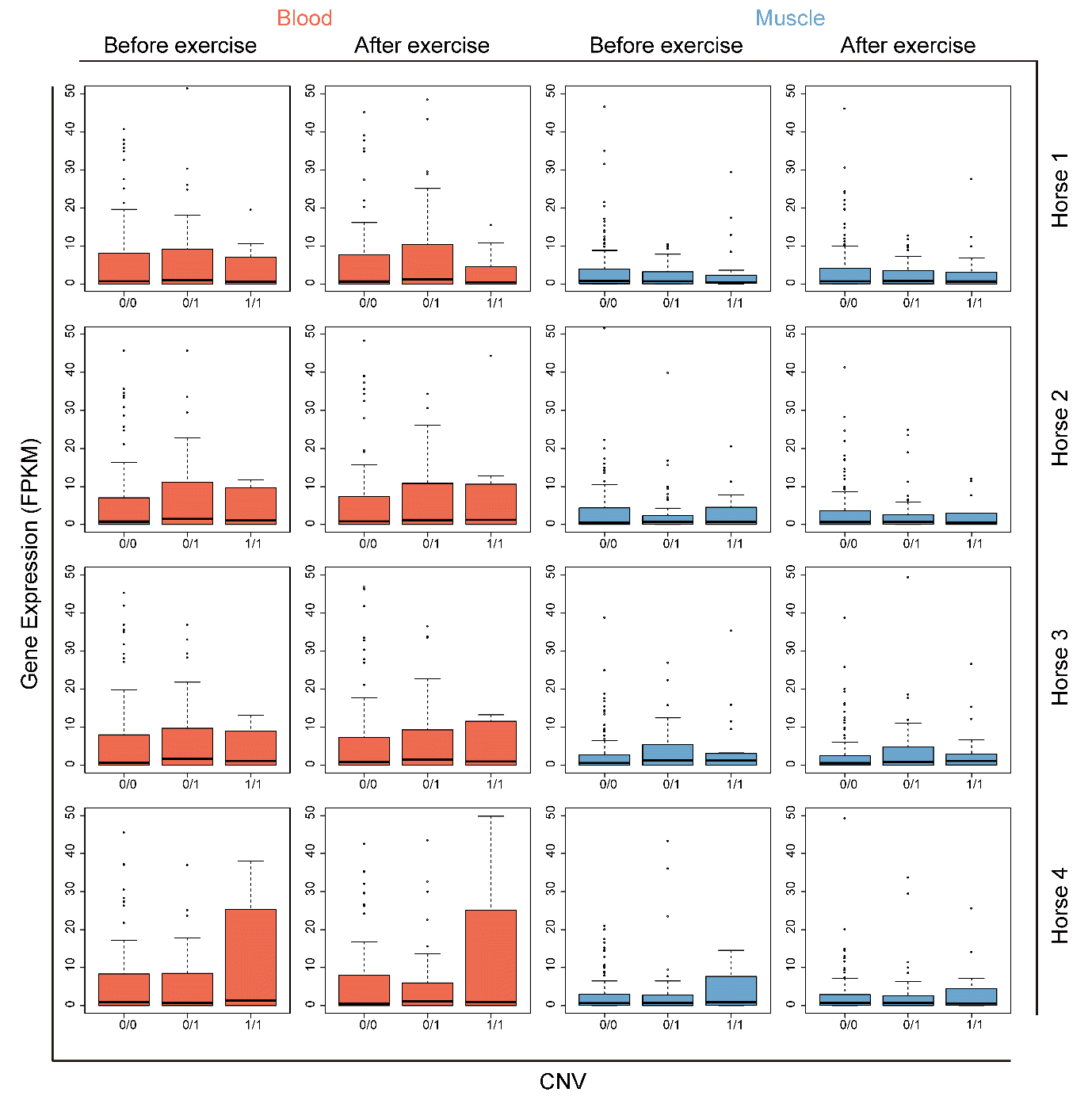
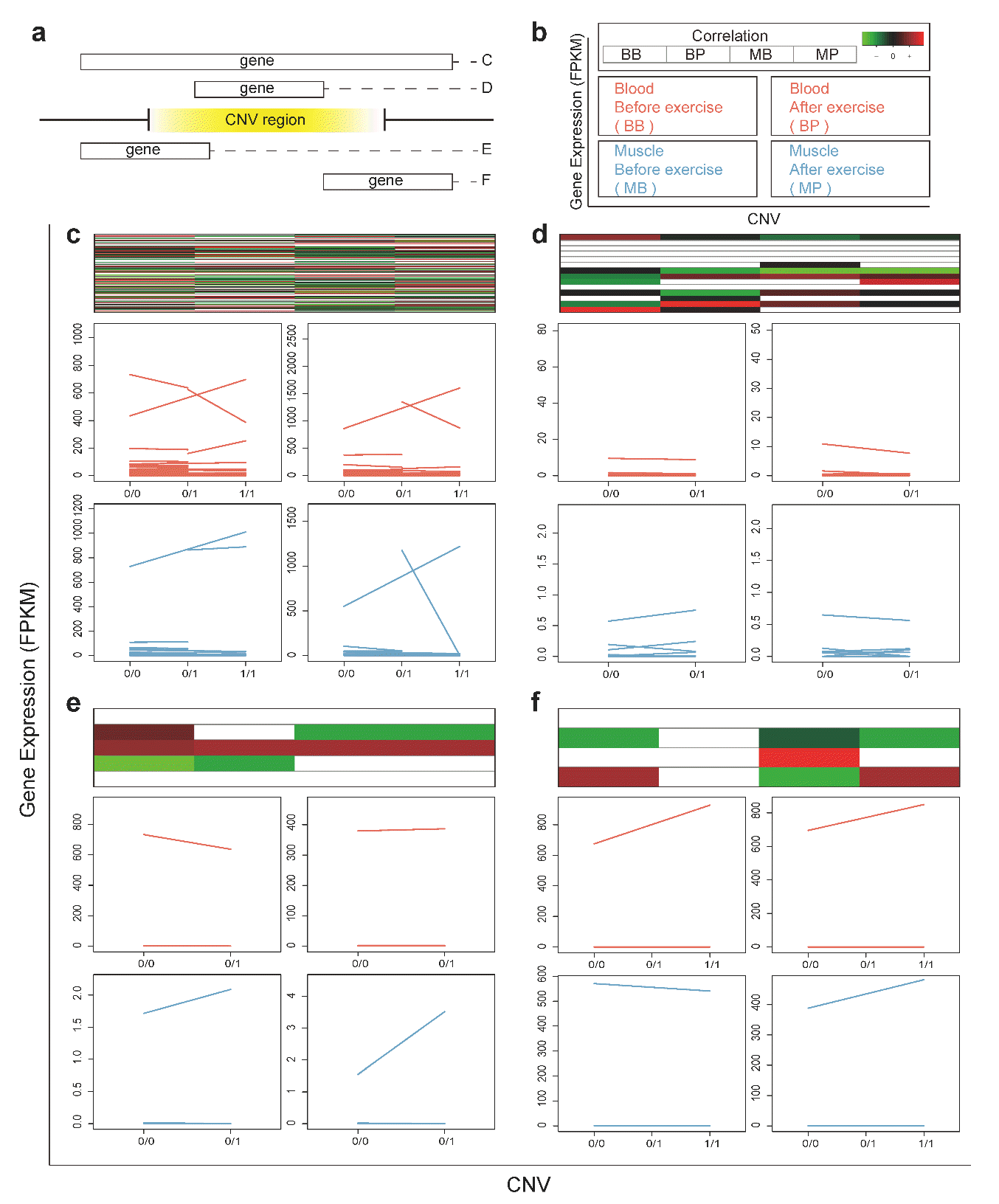
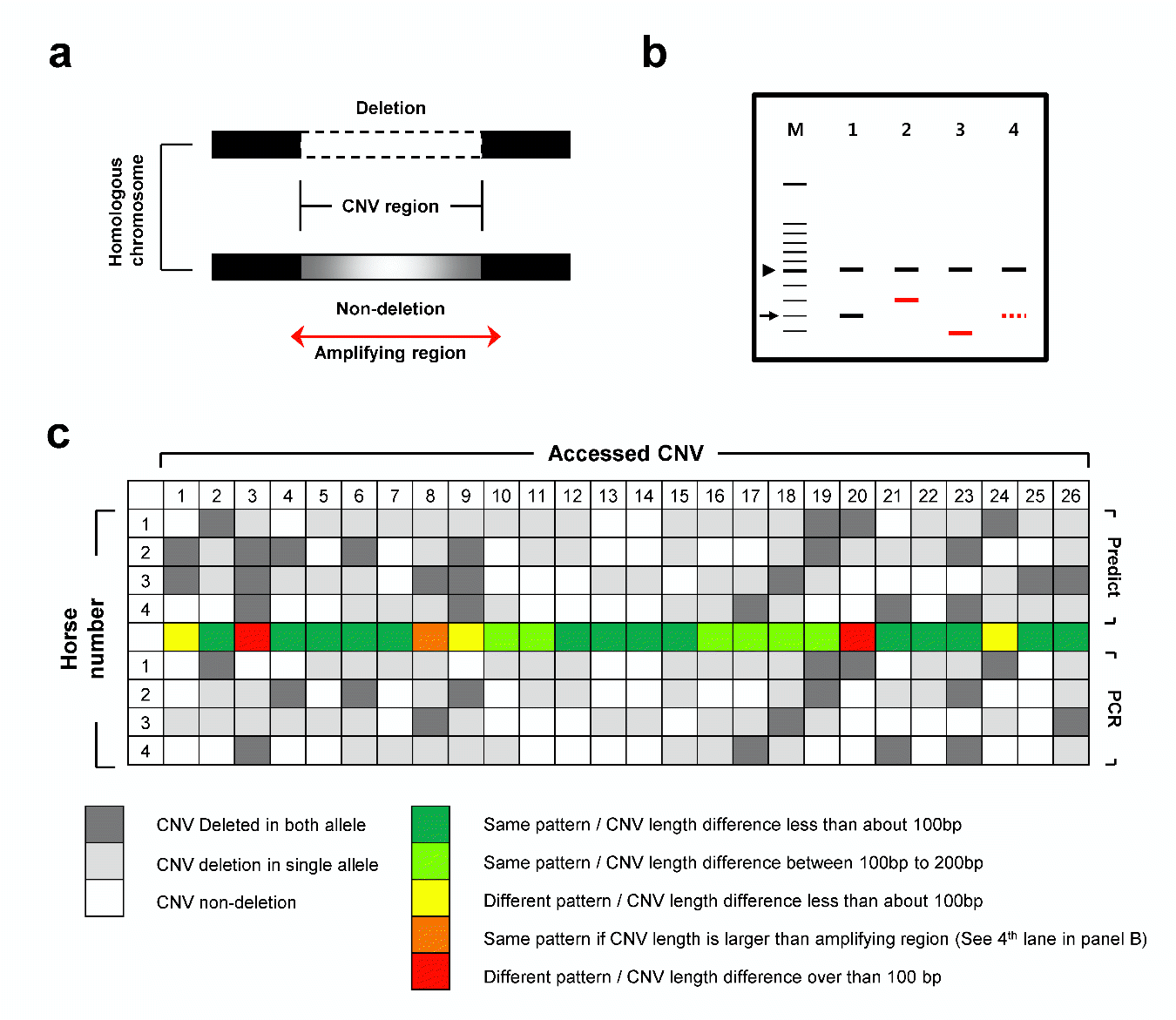
Kim H, Lee T, Park W, Lee JW, Kim J, Lee B-Y, Ahn H, Moon S, Cho S, Do K-T, et al. 2013. Peeling back the evolutionary layers of molecular mechanisms responsive to exercise-stress in the skeletal muscle of the racing horse. DNA Res 20:287ŌĆō298.



Lee JA, Madrid RE, Sperle K, Ritterson CM, Hobson GM, Garbern J, Lupski JR, Inoue K. 2006. Spastic paraplegia type 2 associated with axonal neuropathy and apparent
PLP1 position effect. Ann Neurol 59:398ŌĆō403.


McCarroll SA, Altshuler DM. 2007. Copy-number variation and association studies of human disease. Nat Genet 39:S37ŌĆōS42.


McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297ŌĆō1303.



Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, Abyzov A, Yoon SC, Ye K, Cheetham RK, et al. 2011. Mapping copy number variation by population-scale genome sequencing. Nature 470:59ŌĆō65.



Pankratz N, Dumitriu A, Hetrick KN, Sun M, Latourelle JC, Wilk JB, Halter C, Doheny KF, Gusella JF, Nichols WC, et al. 2011. Copy number variation in familial Parkinson disease. PloS one 6:e20988



Park K-D, Park J, Ko J, Kim BC, Kim H-S, Ahn K, Do K-T, Choi H, Kim H-M, Song S, et al. 2012. Whole transcriptome analyses of six thoroughbred horses before and after exercise using RNA-Seq. BMC Genomics 13:473



Reich DE, Schaffner SF, Daly MJ, McVean G, Mullikin JC, Higgins JM, Richter DJ, Lander ES, Altshuler D. 2002. Human genome sequence variation and the influence of gene history, mutation and recombination. Nature Genet 32:135ŌĆō142.


Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, Sherry S, Mullikin JC, Mortimore BJ, Willey DL, et al. 2001. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409:928ŌĆō933.


Sharp AJ, Cheng Z, Eichler EE. 2006. Structural variation of the human genome. Annu Rev Genomics Hum Genet 7:407ŌĆō442.


Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102:15545ŌĆō15550.



Xi R, Hadjipanayis AG, Luquette LJ, Kim T-M, Lee E, Zhang J, Johnson MD, Muzny DM, Wheeler DA, Gibbs RA, et al. 2011. Copy number variation detection in whole-genome sequencing data using the Bayesian information criterion. Proc Natl Acad Sci 108:E1128ŌĆōE1136.










 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print





