 |
 |
Abstract
Goose fatty liver is one of the most delicious and popular foods in the world, but there is no reliable genetic marker for the early selection and breeding of geese with good liver-producing potential. In our study, one hundred and twenty-four 78-day-old Landes geese bred in Shunda Landes goose breeding farm, Jiutai, Jilin, China were selected randomly. The fatty livers were sampled each week after overfeeding during a three week period. Polymerase chain reaction-single strand conformation polymorphism and DNA sequencing were used to identify single nucleotide polymorphisms (SNPs) of fatty acid synthase (FAS), which is an important enzyme involved in the synthesis of fat under both physiological and pathological conditions. Least-squares correlation was established between these SNPs and fatty liver weight, abdominal fat weight, and intestinal fat weight of the overfed Landes geese, respectively. The results showed that fatty liver weight of geese with EF and FF genotypes (amplified by primer P1) was significantly higher than that of the EE genotype (p<0.05), and liver weight of CD and DD genotypes (amplified by primer P2) was significantly higher than that of the CC genotype (p<0.05). Different genotype combinations showed different liver weights, and from highest to lowest were ABDD, DDEF, DDFF, DDEE, ABEF, ABFF, AADD, and CDEF. Further analysis of DNA sequencing showed that there were two SNPs within the 5ŌĆ▓ promoter region the FAS gene. The geese of EF and FF genotypes carried a change of T to C, and the geese of CD and DD genotypes carried a change of A to G. The changes of the bases could potentially influence the binding of some transcription factors to this region as to regulate FAS gene. To our knowledge, this is the first report of SNPs found within the 5ŌĆ▓ promoter region of the Landes goose FAS gene, and our data will provide an insight for early selection of geese for liver production.
The association between gene polymorphism and a particular trait has been extensively studied in modern animal breeding, and single nucleotide polymorphisms (SNPs) are one type that is extremely useful for marker assisted selection (MAS).
Landes geese are famous for their good fatty liver production, and the average liver weight after overfeeding is about 700 to 800 g. We are interested in the fat deposition in the liver of the overfed goose, a complicated process involving many genes related to the metabolism of carbohydrates, amino acids, and lipids (Zhu et al., 2011). Some reports have shown that SNPs in the genes involved in the fatty acid synthesis may associate with fat deposition. In turkeys, there was a significant association between fatty acid synthase (FAS) gene polymorphisms and fatness variability (Sourdioux et al., 1999). In chicken, gene polymorphisms in fatty acid binding protein strongly correlated with abdominal fat deposition (Wang et al., 2001). Five polymorphic sites in 5ŌĆ▓ and 3ŌĆ▓ regulatory regions of chicken myostatin gene were found to be closely correlated with bone growth and fat metabolism (Gu et al., 2003). But till now, the associations mainly focus on the chicken, there is almost no reports of association between gene polymorphism and liver fat deposition, especially in the fatty liver of the overfed goose.
According to the previous studies, FAS plays important roles in fatty acid synthesis (Shu et al., 2012), and its activity is regulated by insulin signaling and glucose (Han et al., 2009). Consequently, we wanted to explore whether there are SNPs in the FAS gene that were associated with fatty liver weight. In this study, high carbohydrate diet was used to overfeed Landes geese for 3 weeks, at which time the average weight of a fatty liver could reach 800 to 1,000 g, approximately 5 to 10 time than the normal weight. The single-strand conformation polymorphism (SSCP) technique was used to divide the geese into different genotypes. Finally, we found two SNPs in the FAS gene that were significantly associated with fatty liver weight, one was in the promoter region, and the other was in the coding region. The results of our study could provide useful information for geese breeding especially for fatty liver production.
One hundred and twenty-four healthy 78-day-old Landes geese were selected for the overfeeding experiment, which was carried out at the Shunda Landes goose breeding farm, Jiutai, Jilin, China. Each goose was labeled with a number on its wing. Before overfeeding, the average weight of the geese was 4.31┬▒0.25 kg (p>0.05). The overfeeding period lasted for 21 days. Finally, the geese were slaughtered, and the weight of internal and external fat deposition in their livers was determined. The breeding management and experimental procedure of the geese are in conformity with the principles provided by the WMA Declaration of Helsinki.
The parameters determined included the weight before overfeeding, weight at one-week and two-week after overfeeding, weight before slaughter, fatty liver weight, intestinal fat weight, and abdominal fat weight. All of the weights (Mean┬▒standard error) were determined by an electronic scale with an accuracy of ┬▒5 g. Before slaughter, 2 mL blood sample was taken from the wing vein of each goose, and genomic DNA was extracted for further analysis of FAS gene polymorphism (Liu, 2003).
The pre-overfeeding period was one week, during which each goose was fed about 400 g corn every day. When the overfeeding began, the geese were placed in the cages (four geese in each cage). An overfeeding machine was used in our study, and operated by the same person each time. The geese were overfed four times each day at an interval of six hours with 250 to 350 g high carbohydrate diet. The temperature in the goose house was about 20┬░C to 22┬░C, and controlled in as small fluctuation as possible. The relative humidity was 50% to 65%, and internal environment of the house was kept quiet and well ventilated.
Experimental feed formula was composed of 97.6% corn, 0.9% salt, 1% vegetable oil, 0.5% vitamins and minerals. This formula contained 7.8% protein and 14.10 MJ/kg metabolizable energy. The corn was boiled for 30min, and then the other ingredients were added before mixing and stored until use.
According to the chicken FAS gene sequence published in GenBank, nine pairs of primers were designed using Primer Premier 5.0 software, and then synthesized by Shanghai Sangon Biological Engineering Technology and Services Co., Ltd. Finally, the polymorphic variations in the polymerase chain reaction (PCR) products were detected by the sodium dodecyl sulfate polyacrylamide gel electrophoresis technique. The primer sequences of the PCR products with SNPs were shown in Table 1.
The PCR reaction system was composed of 2 ╬╝L 10├ŚPCR buffer, 0.6 ╬╝L 10 ╬╝mol/L dNTPs, 0.6 ╬╝L 10 mmol/L of each primer, 0.5 ╬╝L Taq polymerase, 1.0 ╬╝L DNA template, and 14.7 ╬╝L ddH2O. The amplification conditions were pre-denaturation at 94┬░C for 4min, then 34 cycles of denaturation at 94┬░C for 45 s, annealing at 60┬░C for 45 s, and extension at 72┬░C for 1 min, and finally extension at 72┬░C for 7 min.
The 2 ╬╝L PCR products and 6 ╬╝L loading buffer were mixed, and annealed at 99┬░C for 10 min. Then the mixture was cooled in an ice bath for 5 min, and electrophoresed through 12% non-denaturing polyacrylamide gel (Acrylamide:Bis solution = 29:1; 10 V/cm, 10 h). Finally, the DNA bands were detected by silver staining.
The polymorphic products were recovered and purified by QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany), and then sent to Shanghai Sangon Biological Engineering Technology and Services Co., Ltd. for sequencing.
SPSS version 19 was used for statistical analysis with the following least-squares model.
yij is the fatty liver weight, ╬╝ is the average value, Gi is the fixed effect, eij is the random effect.
This model is used for the single genotype analysis.
yij is the fatty liver weight, ╬╝ is the average value, Gi is the fixed effect for each genotype amplified by the primer P1, P2, and P8, eij is the random effect.
This model is used for the analysis of the genotype combinations.
Hardy-Weinberg test was also calculated (Supplemental data: See e-version for supplement).
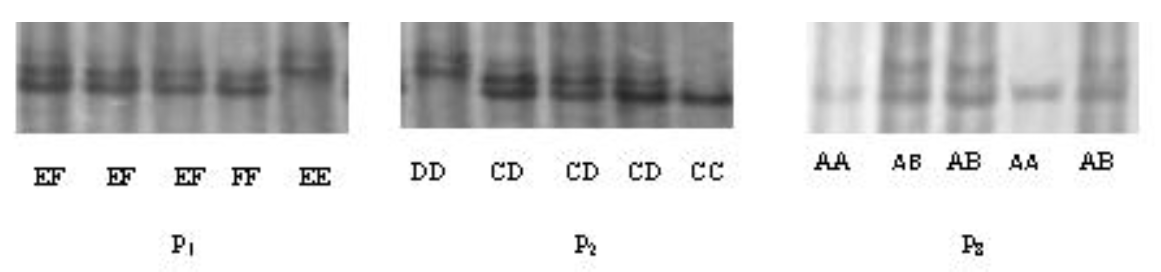
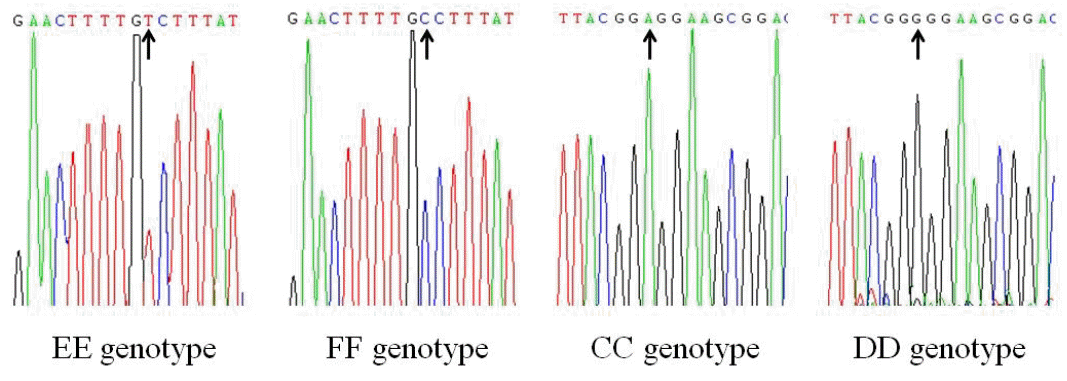
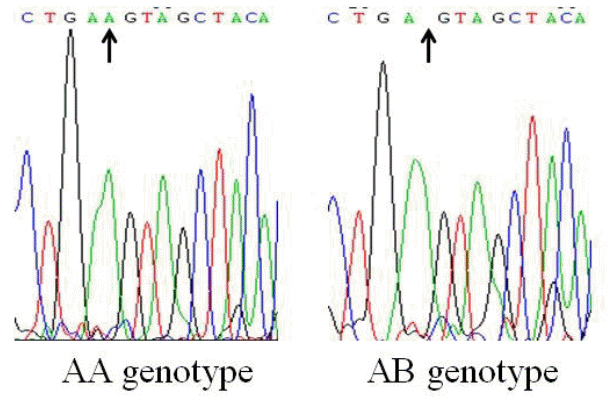
The PCR products amplified by the three of the nine primer pairs had polymorphic sites. According to the electrophoretic patterns, PCR products amplified by primer 1 (P1) were determined as three genotypes, EE, EF, and FF. For primer 2 (P2), three genotypes were CC, CD, and DD, and for primer 8 (P8), there were only two genotypes, AA and AB. The electrophoretic patterns of the primers are shown in Figure 1, and the sequencing results of different genotypes are shown in Figure 2 and 3.
We used TRSEARCH ver.1.3 software to predict the potential regulatory elements and protein-binding sites in the 5ŌĆ▓ promoter region of the goose FAS gene (URL http://www.cbrc.jp/research/DSB/tfsearch.html). The difference in regulation and transcription factor binding at the polymorphic sites before and after the changes of the bases are summarized in Table 2. The changes at these bases altered the predicted transcription factor binding in the 5ŌĆ▓ promoter region in the FAS gene. For polymorphic (T/C) site at position ŌłÆ54, the regulatory factor Oct-1 was predicted to bind when it was T, but when it was changed to C, no regulatory factor was predicted to bind. For polymorphic (A/G) site at position ŌłÆ545, no regulatory factor was predicted to bind when it was A, but when it was changed to G, myeloid zinc finger protein 1 (MZF1) binding was predicted.
The SPSS 19 was used to analyze the relationship between FAS gene SNPs and internal and external fat deposition in the goose fatty liver. The results are shown in Table 3.
The fatty liver weight of EF and FF genotypes was significantly higher than that of the EE-type (p<0.05), so the F genotype favored fat deposition. The fatty liver weight of CD and DD genotypes was significantly higher than that of CC genotype (p<0.05), so the D genotype was favorable. The fatty liver weight of AA genotype was higher than that of AB genotype, but the difference was not significant (p>0.05).
Least-squares analysis on 2├Ś2 and 3├Ś3 multiple genotype combinations was performed to test the relationship between the composite genotypes and fatty liver performance. The results are shown in Table 4 and Supplementary Table S1 (See e-version for supplement).
From Table 4, among the combinations of two genotypes at two sites, the fatty liver weight of the ABDD genotype was the highest, up to 950.23 g, while that of the AAEE genotype was the lowest, 585.23 g. From highest to lowest were ABDD, DDEF, DDFF, DDEE, ABEF, ABFF, AADD, CDEF genotypes (all of which exceeded 800 g). The abdominal fat weight of DDFF and DDEF genotypes was significantly higher than that of AAEE type (p<0.05). The intestinal fat weight of CCFF and AAFF genotypes was significantly higher than that of DDEE type (p<0.05).
The products of P1, P2, and P8 were aligned with the corresponding DNA sequence of FAS in GenBank. A change of base T to C was found at ŌĆō54 bp site in the 5ŌĆ▓ promoter region. The sequence of EE genotype was identical with that of wild type, and FF genotype was a mutant. A change of base A to G was found at ŌĆō545 bp site. The sequence of CC genotype was identical with that of wild type, and DD genotype was a mutant. A single nucleotide deletion of base A was found at 3205 in the coding region. The sequence of AA genotype was identical with that of wild type, and AB genotype was a mutant.
TATA box and CAAT box were found between ŌĆō60 bp and ŌĆō28 bp at the 5ŌĆ▓ end of the goose FAS gene (Kameda, 1991). They found that this region was very important for gene transcription and expression, as the mutation of several nucleotides or even one nucleotide led to changes in transcription. In our study, two SNPs were found in the 5ŌĆ▓ promoter region. One was T to C mutation at ŌĆō54 bp, close to the transcriptional start site and the TATA box, which could function as an important transcriptional element. TRSEARCH ver.1.3 software was used to predict the effects of this change on potential transcriptional regulatory elements and protein binding sites, and we found that when a T was present at ŌĆō54 bp, Oct-1 binding was predicted, while when it was C, no regulatory factor was predicted to bind. According to the correlation analysis, this mutation had a significant effect on fatty liver weight, consistent with KamedaŌĆÖs study. Oct-1 is a member of the pit-oct-unc transcription factor family, and expresses in most cells, and regulates the expression of a variety of housekeeping genes. Lack of Oct-1 may affect the expression and regulation of FAS, but further study is needed to uncover the specific mechanism.
The other was A to G mutation at ŌĆō545 bp. When base A was present at this site, no regulatory factors were predicted to bind, but when the base was changed to G, MZF1 binding was predicted. The MZF-1 binding sites are present in the promoters of several genes. It can achieve transcriptional regulation through binding DNA or RNA, or directly regulate gene transcription by forming complexes with zinc finger proteins. Lack of MZF1 activity greatly increased cell differentiation in mice (Gaboli et al., 2001). Here we found that the presence of the MZF1 binding site might significantly affect FAS transcription and fatty liver weight. According to the strong correlation of the SNPs and the fatty liver weight, we concluded that these two sites could be used as candidates for MAS of the fatty liver-producing trait.
A study on commercial turkey breeding showed that a restriction enzyme site of MspI within FAS correlated with fat deposition (Sourdioux et al., 1996). Four hundred and ten Microsatellite markers on 25 chicken chromosomes were screened for quantitative trait loci (QTLs), and found that there were multiple QTLs within FAS gene (Jennen et al., 2004). So, the SNPs we found could locate in QTLs which control the fat deposition in the liver.
In recent years, research on the correlation of human diseases with genetic variation has shown that correlation analysis of combining multiple SNPs with the trait is much more precise than that of single SNP. Here, the correlation analysis between the combined genotypes of multiple FAS gene sites and the fatty liver trait showed that the combination of two genotypes among three polymorphic sites made a significant difference in the fatty liver weight (p<0.05). The ABDD genotype was the strongest combination of all genotypes, and the average fatty liver weight reached 950.23 g, which was significantly different from the weight of the other genotypes (p<0.05). The effect of the single-site genotype of P8 on the fatty liver weight was not significant, but the combination ABDD genotype was significantly different from other combination genotypes, and this may reflect the synergistic effect of P1 and P8. From Table 4, we can see that the genotypes with higher fatty liver weight were mostly combination of D and F alleles, which suggests that these two alleles could function synergistically. The correlation analysis between the three-site combination genotypes and the fatty liver weight revealed that the combination genotypes did not show significant differences in fatty liver traits, which may due to the small sample number. But the genotypes with larger fatty livers all had D and F alleles, further confirming that there was synergy between them. The statistical analysis of fatty liver weight with different genotype combination showed that B, D, and F were the dominant alleles, because the fatty liver weight of their heterozygous or homozygous genotypes were significantly or very significantly higher than those of other homozygous or heterozygous genotypes (p<0.05 or p<0.01).
The average fatty liver weight of genotypes without any of the three advantageous alleles B, D, or F was 516.70┬▒100.10 g. The average fatty liver weight of combination genotypes containing one dominant allele was 688.50┬▒174.91 g, which was 71.82 g higher than that of other combination genotypes containing no advantageous alleles. The average fatty liver weight of combination genotypes containing two dominant alleles was 721.80┬▒171.67 g, which was 33.39 g higher than that of combination genotypes having one dominant allele. The average fatty liver weight of combination genotypes containing three dominant alleles was 827.58┬▒194.64 g, which was 105.69 g higher than that of combination genotypes containing two advantageous alleles (p<0.01). The average fatty liver weight of combination genotypes having four advantageous alleles was 64.92 g higher than that of those containing three advantageous alleles (p<0.05) and was extremely significantly (375.80 g) higher than that of the combination genotypes containing no advantageous alleles. All the above results fully proved that the D, F, and B alleles are the dominant alleles for goose fatty liver weight, though due to the small sample number. Further large scale experiments will be required to confirm the synergy between the three alleles.
The results also showed that the combination genotypes not only affected fatty liver weight, but also affected abdominal and intestinal fat weight. Among the single genotypes, there was no significant difference for each trait, but in combination genotypes of two sites, difference in each trait between different genotypes was significant (p<0.05). It suggested that different sites might exert minor cumulative effect. The body fat level at different growth periods are positively correlated with liver FAS activity (Tian et al., 1996). The results of this study are consistent those results.
In conclusion, our study identified two SNPs in the 5ŌĆ▓ promoter region of Landes goose FAS gene, and correlation analysis showed for the first time that these SNPs associated with fatty liver weight. The alleles D, F, and B were dominant for goose fatty liver weight, and they could be used in MAS for the fatty liver trait.
Figure┬Ā1
Band patterns of the polymorphic polymerase chain reaction products amplified by three pairs of primers.
Figure┬Ā2
Single nucleotide polymorphisms of EE, FF, CC, and DD genotypes, polymerase chain reaction products of primer 1 and 2.
Figure┬Ā3
Single nucleotide polymorphisms of AA and AB genotypes, polymerase chain reaction products of primer 8.
Table┬Ā1
Primer sequences for amplification of the FAS gene
Table┬Ā2
Prediction for the binding site of transcription factors with the 5ŌĆ▓ region of FAS gene
| Position of the binding site | Mutation type | Transcription factor | Transcription factor |
|---|---|---|---|
| Site 1 ( 54 bp) | TŌåÆC | Oct-1 | - |
| Site 2 ( 545 bp) | AŌåÆG | - | MZF1 |
Table┬Ā3
Relationship between genotypes and the performance of geese
Table┬Ā4
Relationship between different genotype combinations and the performance of geese
REFERENCES
Gaboli M, Kotsi PA, Gurrieri C, Cattoretti G, Ronchetti S, Cordon-Cardo C, Broxmeyer HE, Hromas R, Pandolfi PP. 2001. Mzf1 controls cell proliferation and tumorigenesis. Genes Dev 15:1625ŌĆō1630.



Gu ZL, Zhu DH, Li N, Li H, Deng XM, Wu CX. 2003. Association of myostatin SNPs with growth of muscle and fatness. Sci China C Life Sci 33:273ŌĆō280.
Han CC, Wang JW, Li L, Zhang ZX, Wang L, Pan ZX. 2009. The role of insulin and glucose in goose primary hepatocyte triglyceride accumulation. J Exp Biol 212:1553ŌĆō1558.


Jennen DG, Vereijken AL, Bovenhuis H, Crooijmans RP, Veenendaal A, van der Poel JJ, Groenen MA. 2004. Detection and localization of quantitative trait loci affecting fatness in broilers. Poult Sci 83:3295ŌĆō301.


Kameda K, Goodridge AG. 1991. Isolation and partial characterization of the gene for goose fatty acid synthase. J Biol Chem 266:419ŌĆō426.


Liu CG. 2003. Preservation of poultry blood samples and its DNA extraction. Acta Agriculturae Boreali-occidentalis Sinica 12:33ŌĆō35.
Shu CP, Wang BW, Li Z, Ge WH, Zhang MA, Yue B. 2012. Fatty acids deposition and FAS mRNA expression abundance in liver tissue of overfeeding goose. China Agric Sci 45:2002ŌĆō2011.
Sourdioux M, Douaire M, Delabrosse Y. 1996. DNA polymorphisms of lipogenesis genes and analysis of linkage with fatness in turkeys. Poult Sci 75:1018ŌĆō1026.


Sourdioux M, Brevelet C, Delabrosse Y, Douaire M. 1999. Association of fatty acid synthase gene and malic enzyme gene polymorphism with fatness in turkey. Poult Sci 78:1651ŌĆō1657.


Tian WX, Dong Y, Quan H, Chen WF. 1996. The dependence of fat level of hen on activity of fatty acid synthase in liver on different ages. Chinese Biochem J 12:324ŌĆō326.
Wang QG, Li N, Deng XM, Lian ZX, Li H, Wu CX. 2001. Relationship of extracellular fatty acid binding protein SNPs with abdominal fatness in chicken. Sci China C Life Sci 31:266ŌĆō270.





 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print





