 |
 |
Abstract
As a member of a subclass of immunophilins, it is controversial that FKBP38 acts an upstream regulator of mTOR signaling pathway, which control the process of cell-growth, proliferation and differentiation. In order to explore the relationship between FKBP38 and mTOR in the Cashmere goat (Capra hircus) cells, a full-length cDNA was cloned (GenBank accession number JF714970) and expression pattern was analyzed. The cloned FKBP38 gene is 1,248 bp in length, containing an open reading frame (ORF) from nucleotide 13 to 1,248 which encodes 411 amino acids, and 12 nucleotides in front of the initiation codon. The full cDNA sequence shares 98% identity with cattle, 94% with horse and 90% with human. The putative amino acid sequence shows the higher homology which is 98%, 97% and 94%, correspondingly. The bioinformatics analysis showed that FKBP38 contained a FKBP_C domain, two TPR domains and a TM domain. Psite analysis suggested that the ORF encoding protein contained a leucine-zipper pattern and a Prenyl group binding site (CAAX box). Tissue-specific expression analysis was performed by semi-quantitative RT-PCR and showed that the FKBP38 expression was detected in all the tested tissues and the highest level of mRNA accumulation was detected in testis, suggesting that FKBP38 plays an important role in goat cells.
A new member of FKBP (FK506-binding protein) family, FKBP38, was isolated from a cDNA library of human T cells in the early 1990s (Lam et al., 1995). Then, some researchers showed that FKBP38 gene was located on chromosome 19, area p12 in human, and actually, that the FKBP38 is a truncated product of FKBP8 gene insisting of 9 exons (Nielson et al., 2004). FKBP38 contains three kinds of domain, FKBP_C domain, TPR domain, and TM domain. In the absence of growth factors and nutrition, FKBP_C domain may bond to the mammalian target of rapamycin (mTOR) to restrain the phosphorylation of S6K1 and 4EBP1 (Bai et al., 2007) which are the downstream regulatory factors of mTOR signaling pathway. While in conditions of rich growth factors and nutrients, FKBP_C domain may interact with Rheb (Ras homolog enriched in brain)-GTP to activate the mTOR downstream signaling pathway by releasing mTOR from FKBP38 (Bai et al., 2007). With the interaction between TPR domain of FKBP38 and S4 subunit of 26S proteasome, FKBP38 could anchor the 26s proteasome at the organelle membrane (Nakagawa et al., 2007). With the help of TM domain, FKBP38 can locate Bcl-2 and Bcl-Xl at the mitochondrial membrane, which are key regulators of apoptosis pathway (Kang et al., 2005; Wang et al., 2005; Chen et al., 2008). All experimented mice, with knockout FKBP38 gene, would have different congenital malformations (Bulgakov et al., 2004; Shirane et al., 2008; Wong et al., 2008). This demonstrates that FKPB38 plays a key role in early embryonic development.
There is some evidence indicating that Rheb, a small guanosine triphosphate (GTP)-binding protein, interacts with mTOR and activates the enzymatic activity of mTOR by binding the Rheb-GTP to mTOR directly (Long et al., 2005; Long et al., 2007). However, the functional relationship between Rheb and mTOR is puzzling because of the identification of FKBP38, a mitochondrial membrane protein, as an endogenous mTOR inhibitor. Rheb-GTP can apparently induce the releasing of mTOR from FKBP38 through interacting with FKBP38 directly (Bai et al., 2007; Ma et al., 2008). In this model, GTP-bound Rheb binds to FKBP38 and induces the release and activation of mTOR. However, a later paper reports that the interaction between Rheb and FKBP38 could not be detected in three different in vitro assays (Uhlenbrock et al., 2009). The detailed mechanism of the interaction between Rheb and FKBP38 requires further investigation as the link between mTOR, Rheb and FKBP38 is not clear.
Although the FKBP38 has been identified in mouse (NP_034353) and human (AAO39020), its physiological function has not been fully investigated and remains to be unclearly defined. In the present study, in order to explore the function of FKBP38 in goat cells, we cloned the code sequence fragment of the FKBP38 gene cDNA from Inner Mongolia Cashmere goat (Capra hircus) and analyzed its molecular characterization using the basic bioinformatics methods. Semi-quantitative RT-PCR was used to detect the relative expression levels in various tissues of the Cashmere goat.
Inner Mongolia Cashmere goats were bred on a natural diet in Inner Mongolia, China. Tissues including testis, brain, liver, lung, mammary gland, spleen and kidney were collected from the goats after slaughter. Tissue samples were immediately frozen in liquid nitrogen and stored at ŌłÆ80┬░C.
Inner Mongolia Cashmere goat fetal fibroblasts (GFb cells) were maintained as monolayer cultures in DMEM/F12 (D-MEM/F-12, Gibco, Paisley, PA49RF, Scotland, UK) supplemented with 10% fetal bovine serum (FBS) and 100 U/ml penicillin G and 100 mg/ml1 streptomycin (FBS, Hyclone Laboratories, Inc. Logan, UT USA and penicillin/streptomycin, Sigma-Aldrich, Inc. St. Louis USA). Cell cultures were maintained and incubated at 37┬░C in humidified air with 5% CO2.
Total RNA was isolated from testis, brain, liver, lung, mammary gland, spleen, kidney and fetal fibroblast cells of Inner Mongolia goats, using an RNAzol kit (RNAiso Plus, TaKaRa Co. Ltd., Dalian, China). RNA concentration was determined spectrophotometrically (J231, Thermo Fisher, Waltham, USA). mRNAs were reversely transcribed with an AMV 1st strand cDNA Synthesis kit and an oligo d (T) 18 primer according to manufacturerŌĆÖs protocol (Takara Co. Ltd, Dalian, China).
To amplify FKBP38 gene code sequence (CDS) fragment from the Cashmere goat, a pair of specific primers were designed (fw: 5ŌĆ▓-GATCCCAGCAGCATGGCGTCT -3ŌĆ▓; brv: 5ŌĆ▓-TCAGTTCCTGGCAGCAATGAC -3ŌĆ▓) based on the human (AY278607), cattle (BC122594) and mouse (AY225340) FKBP38 gene nucleotide sequence. PCR was conducted with an initial denature action step at 94┬░C for 4 min, followed by 94┬░C for 30 s, 70┬░C for 30 s, and 72┬░C for 90s, 35cycles and 72┬░C for 10 min. PCR reaction mixture (10 ╬╝l) containing TaKaRa LA Taq (TaKaRa Taq Version, Takara Co. Ltd., China) 0.1 ╬╝l, dNTP Mixture (2.5 mm each) 1.6 ╬╝l, template cDNA 1.0 ╬╝l, fw (10 ╬╝M) and rv (10 ╬╝M) mixture 2.0 ╬╝l, 10├Ś LA PCR Buffer, (Mg2+ plus) 1 ╬╝l and d3H2O 4.3 ╬╝l. PCR products were electrophoresed and analysis made by an electronic UV transilluminator (UVItec, London, UK). The PCR products were purified and cloned into a pMD19-T vector (Takara Co. Ltd, Dalian, China) followed by sequencing.
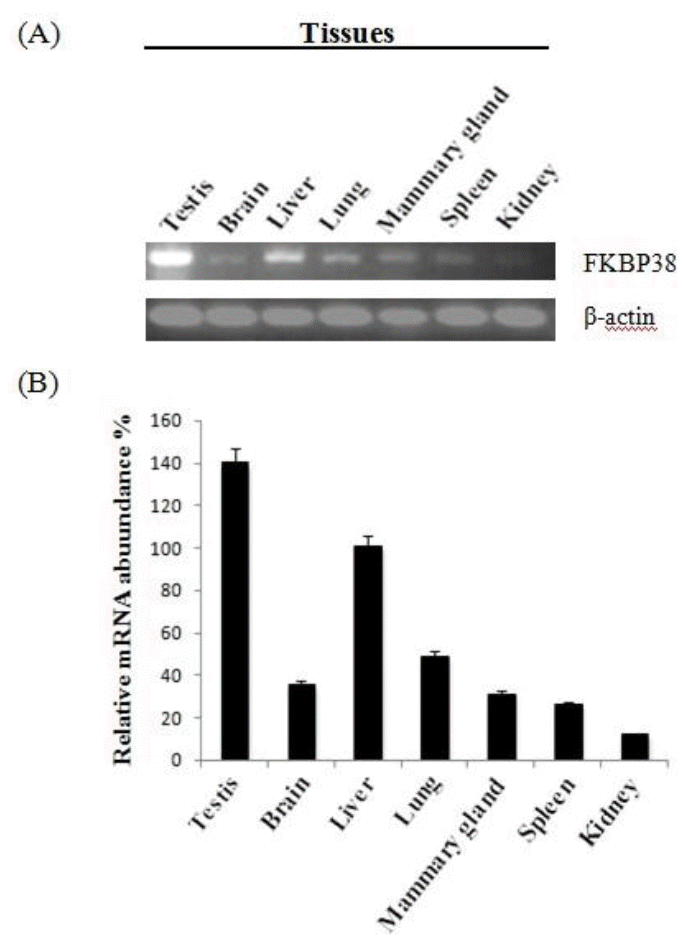
Tissue distribution of FKBP38 mRNA was performed using semi-quantitative RT-PCR analysis. Total RNA from testis, brain, liver, lung, mammary gland, spleen and kidney was extracted and converted to cDNA. The PCR amplifications were performed in 10 ╬╝l total volume for 30 cycles at the appropriate annealing temperature with the primers similar to that of the CDS fragment. FKBP38 mRNA was detected in different tissues while ╬▓-actin as a loading control.
Nucleotide sequences of goat FKBP38 cDNA and deduced amino acid sequence was accomplished by the NCBI BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/). Predictions of open reading frames (ORFs) and theoretical molecular weights of deduced polypeptides were performed by the Protein property calculator (http://www.basic.northwestern.edu/biotools/proteincalc.html). The protein Isoelectric Point was predicted by the calculation of protein isoelectric point (http://isoelectric.ovh.org/). Subcellular localization of the FKBP38 was predicted by the PSORT program (http://psort.ims.u-tokyo.ac.jp/form2.html). Protein domain analysis was searched by the SMART program (http://smart.embl-heidelberg.de/) and the EMBL-EBI InterProScan program (http://www.ebi.ac.uk/Tools/pfa/iprscan/). Protein prosite patterns analysis was identified by the Psite program (http://www.softberry.com). The bands on gel were analyzed by Carestream MI software (http://www.carestream.com/). A phylogenetic tree was constructed by MEGA4.1 (http://www.megasoftware.net/).
The cDNA of FKBP38 gene (GenBank accession number JF714970) from Inner Mongolia Cashmere goat was amplified by RT-PCR. The cloned cDNA fragment was 1,248 bp in length and analysis of the sequence revealed the ORF from nucleotide 13 to 1,248 encoding deduced 411 amino acid residues. The full cDNA nucleotide sequence shares 98%, 94%, 90% identity with cattle, horse, and human, respectively. The putative amino acid sequence shows the high homology which is 98%, 97% and 94%, correspondingly.
To elucidate phylogenetic relationships of FKBP38, the amino acid sequence was aligned with other homologous animal FKBP38. Phylogenetic tree based on protein sequences was constructed as shown in Figure 1.
The deduced FKBP38 protein of the Cashmere goat consists of 411 amino acid residues and its predicted molecular weight is 44,404 Da for the unmodified protein and the estimated isoelectric point (pI) is 4.53. The basic amino acids comprise 12.4% Leu, 11.7% Ala, 10.0% Glu, 8.8% Pro, 6.7% Val and 6.7% Gly. The putative FKBP38 protein contains a FKBP_C domain starting at position 114 and ending at position 199, two TPR domains from amino acid 271 to 304 and amino acid 305 to 338, and a TM domain from the position 389 to 408 (Figure 2). There are 2 N-glycosylation sites, 6 protein kinase C phosphorylation sites, 7 Casein kinase II phosphorylation sites, 7 Microbodies C-terminal targeting signals, 1 cAMP- and cGMP-dependent protein kinase phosphorylation site, 1 Tyrosine kinase phosphorylation site, 1 Prenyl group binding site (CAAX box), and 1 Leucine zipper pattern within the FKBP38 protein. The protein prosite comparison of FKBP38 with that of other animals was constructed (Figure 3). Its predicted subcellar location is in mitochondria.
To determine the steady-state level expression of FKBP38 gene in different tissues of goat, semi-quantitative RT-PCR analysis was carried out with total RNA extracted from testis, brain, liver, lung, mammary gland, spleen, kidney (Figure 4). FKBP38 gene was expressed in all the tested tissues and the highest level of mRNA accumulation was detected in testis.
As a member of FKBPs family, FKBP38 was thought to have PPIase (peptidyl-prolyl cis-trans isomerase) activity. However, some researchers reported that unlike other characterized members of the FKBPs family, FKBP38 may not possess detectable PPIase activity because the FKBP_C domain in FKBP38 contains a number of mutations at the PPIase active site (Lam et al., 1995; Liu, 2003; Weiwad et al., 2005). As shown in Figure 2, FKBP38 contains a leucine zipper motif from amino acid 277 to 298 which overlaps the first TRP domain, indicating that FKBP38 may form multimers with itself through interaction with other leucine-zipper-containing, or coiled-coiled-containing proteins (Lam et al., 1995). In addition, mutants without the TM domain were unable to associate with mitochondria the result of which was that FKBP38 lost the ability to inhibit mTOR, suggesting that mitochondrial localization is essential for FKBP38 having an inhibitory activity on the mTOR signaling pathway (Ma et al., 2008). Meanwhile, it seems that the CAAX box has some relationships to mitochondrial localization (Shirane and Nakayama, 2003). All indicated features in predicted domains (Figure 2) and in predicted active sites (Figure 3) are very similar to previous research results of FKBP38 in other species. These results indicated that FKBP38 gene was correctly cloned from Inner Mongolia Cashmere goat.
Rheb is an upstream regulatory factory in mTOR signaling pathway and the Rheb-GTP can activate mTOR to advance the signals to endogenous S6K1 by phosphorylation at its residues Thr389, Thr421 and Ser 424 (Castro et al., 2003; Sarbassov et al., 2005; Sato et al., 2008). Rheb contains a RAS domain, including switch I region and switch II region. It has been demonstrated that the switch I region can interact with the FKBP_C domain of FKBP38 to remove it from mTORC1 in cells (Maestre-Mart├Łnez et al., 2006; Ma et al., 2008). Meanwhile, some evidence showed that the FKBP38 protein lacking the FKBP_C domain, which was highly similar to FKBP12, was unable to interact with Rheb (Ma et al., 2008). As shown above, goat FKBP38 has a FKBP_C domain. Meanwhile, we have cloned the goat Rheb gene and a Ras family conservative domain was predicted from the deduced amino acid sequence (GenBank accession number HM569224). Based on this information, we propose a hypothesis: The FKBP_C domain of FKBP38 and the switch I region of Rheb may interact with each other and regulate the mTOR signaling pathway in goat cells. However, the exact mechanism still needs to be further studied.
The results of semi-quantitative RT-PCR in adult goat tissues indicated that FKBP38 was expressed in various tissues. It was reported that FKBP38 expression was enriched in brain but restricted to the heart, placenta, lung and skeletal muscle tissues in human (Lam et al., 1995). In this study, the goat FKBP38 mRNA was expressed at the comparatively highest level in testis and strongly expressed in liver while its expression was restricted in brain, lung, mammary gland, spleen and kidney. These results may suggest that FKBP38 play a more important role in testis and liver than in low expression level tissues. So its function in goat may be different from that in human.
In conclusion, our data shows that FKBP38 cDNA was cloned from the Inner Mongolia Cashmere goat. The cloned gene is 1,248 bp in length, including a complete ORF corresponding to a polypeptide of 411 amino acids and 12 nucleotides in front of the initiation codon. FKBP38 gene was expressed in all the tested tissues and the highest level of mRNA accumulation was detected in testis tissue.
ACKNOWLEDGEMENTS
This work supported by grant from Natural Sciences Foundation of China (31160469) and Natural Sciences Foundation of Inner Mongolia (2011MS0521), China, and graduate student research projects of Inner Mongolia University and Major Projects for New Varieties of Genetically Modified Organisms (2011ZX08008-002), China.
Figure┬Ā1
Phylogenetic tree for FKBP38 protein in seven species. The deduced goat FKBP38 amino acid sequence was aligned with other homologous animal FKBP38. The phylogenetic tree was constructed by neighbor-joining method using MEGA4.1 software. The species and gene GenBank accession number of FKBP38 protein are Bos taurus (NM_001205721), Canis lupus familiaris (XM_541935), Equus caballus (XM_001503329), Homo sapiens (L37033), Mus musculus (AY225340) and Rattus norvegicus (BC107454).

Figure┬Ā2
The predicted domain of goat FKBP38 protein. One FKBP_C domain locates from amino acid 114 to 199. Two TPR domains locate from amino acid 271 to 304 and amino acid 305 to 338. And one TM domain locates from amino acid 389 to 408. The FKBP_C domain and TPR domain were predicted with EMBL-EBI InterProScan program (http://www.ebi.ac.uk/Tools/pfa/iprscan/) and the TM domain was predicted with SMART Software (http://smart.embl.de/).

Figure┬Ā3
Predicted Psites of FKBP38. Alignment of the amino acid sequence of capra hircus (JF714970), bos taurus (NP_001192650), homo sapiens (AAO39020), rattus norvegicus (NP_001032257), mus musculus (NP_034353). Predicted N-glycolsylation site is marked by pane. Protein kinase C phosphorylation sites are marked by double underlines. Casein kinase II phosphorylation sites are indicated by double wavy lines. Prenyl group binding site (CAAX box) is marked by gray shadow. Microbodies C-terminal targeting signal sites are marked by black shadow. cAMP- and cGMP-dependent protein kinase phosphorylation sites are marked by wavy line. Tyrosine kinase phosphorylation site are marked by a hidden line. Leucine zipper pattern are marked by an underline. All these were determined using the Psite software (http://www.softberry.com).

REFERENCES
Bai X, Ma D, Liu A, Shen X, Wang QJ, Liu Y, Jiang Y. 2007. Rheb activates mTOR by antagonizing its endogenous inhibitor, FKBP38. Science 318:977ŌĆō980.


Bulgakov OV, Eggenschwiler JT, Hong DH, Anderson KV, Li T. 2004. FKBP8 is a negative regulator of mouse sonic hedgehog signaling in neural tissues. Development 131:2149ŌĆō2159.


Castro AF, Rebhun JF, Clark GJ, Quilliam LA. 2003. Rheb binds tuberous sclerosis complex 2 (TSC2) and promotes S6 kinase activation in a rapamycin-and farnesylation-dependent manner. J Biol Chem 278:32493ŌĆō32496.


Chen Y, Sternberg P, Cai J. 2008. Characterization of a Bcl-XL-interacting protein FKBP8 and its splice variant in human RPE cells. Invest Ophthalmol Vis Sci 49:1721ŌĆō1727.



Kang CB, Feng L, Chia J, Yoon HS. 2005. Molecular characterization of FK-506 binding protein 38 and its potential regulatory role on the anti-apoptotic protein Bcl-2. Biochem Biophys Res Commun 337:30ŌĆō38.


Lam E, Martin M, Wiederrecht G. 1995. Isolation of a cDNA encoding a novel human FK506-binding protein homolog containing leucine zipper and tetratricopeptide repeat motifs. Gene 160:297ŌĆō302.


Liu JO. 2003. Endogenous protein inhibitors of calcineurin. Biochem Biophys Res Commun 311:1103ŌĆō1109.


Long X, Lin Y, Ortiz-Vega S, Busch S, Avruch J. 2007. The Rheb switch 2 segment is critical for signaling to target of rapamycin complex 1. J Biol Chem 282:18542ŌĆō18551.



Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. 2005. Rheb binds and regulates the mTOR kinase. Curr Biol 15:702ŌĆō713.


Ma D, Bai X, Guo S, Jiang Y. 2008. The switch I region of Rheb is critical for its interaction with FKBP38. J Biol Chem 283:25963ŌĆō25970.



Maestre-Mart├Łnez M, Edlich F, Jarczowski F, Weiwad M, Fischer G, L├╝cke C. 2006. Solution structure of the FK506-binding domain of human FKBP38. J. Biomol NMR 34:197ŌĆō202.


Nakagawa T, Shirane M, Iemura S, Natsume T, Nakayama KI. 2007. Anchoring of the 26S proteasome to the organellar membrane by FKBP38. Genes Cells 12:709ŌĆō719.


Nielsen JV, Mitchelmore C, Pedersen KM, Kjaerulff KM, Finsen B, Jensen NA. 2004. Fkbp8: novel isoforms, genomic organization, and characterization of a forebrain promoter in transgenic mice. Genomics 83:181ŌĆō192.


Sarbassov D, Ali SM, Sabatini DM. 2005. Growing roles for the mTOR pathway. Curr Opin Cell Biol 17:596ŌĆō603.


Sato T, Umetsu A, Tamanoi F. 2008. Characterization of the Rheb-mTOR signaling pathway in mammalian cells: constitutive active mutants of Rheb and mTOR. Methods Enzymol 438:307ŌĆō320.



Shirane M, Nakayama KI. 2003. Inherent calcineurin inhibitor FKBP38 targets Bcl-2 to mitochondria and inhibits apoptosis. Nat Cell Biol 5:28ŌĆō37.


Shirane M, Ogawa MM, Motoyama J, Nakayama KI. 2008. Regulation of apoptosis and neurite extension by FKBP38 is required for neural tube formation in the mouse. Genes Cells 13:635ŌĆō651.


Uhlenbrock K, Weiwad M, Wetzker R, Fischer G, Wittinghofer A, Rubio I. 2009. Reassessment of the role of FKBP38 in the Rheb/mTORC1 pathway. FEBS Lett 583:965ŌĆō970.


Wang HQ, Nakaya Y, Du Z, Yamane T, Shirane M, Kudo T, Takeda M, Takebayashi K, Noda Y, Nakayama KI. 2005. Interaction of presenilins with FKBP38 promotes apoptosis by reducing mitochondrial Bcl-2. Hum Mol Genet 14:1889ŌĆō1902.







 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print





