2. Loomis PR, Graham JK. Commercial semen freezing: individual male variation in cryosurvival and the response of stallion sperm to customized freezing protocols. Anim Reprod Sci 2008; 105:119–28.
https://doi.org/10.1016/j.anireprosci.2007.11.010


3. Hoffmann N, Oldenhof H, Morandini C, Rohn K, Sieme H. Optimal concentrations of cryoprotective agents for semen from stallions that are classified ‘good’ or ‘poor’ for freezing. Anim Reprod Sci 2011; 125:112–8.
https://doi.org/10.1016/j.anireprosci.2011.03.001


5. Sakhatsky NI, Tereshchenko AV, Artemenko AB. An express-method of estimation of fertilizing ability of freezing-thawing of poultry spermatozoa. Sel’skokhozyaistvennaya Biol 1987; 22:77–80.
7. Mel’nyk YuF, Mykytyuk DM, Bilous OV, et al. Program of preservation of the gene pool of main types of farm animals in Ukraine for the period till 2015. Kyiv, Ukraine: Aristey; 2009.
8. Weigend S, Romanov MN, Rath D. Methodologies to identify, evaluate and conserve poultry genetic resources. In : XXII World’s Poultry Congress & Exhibition: Participant List & Full Text CD + Book of Abstracts; 2004 Jun 8–13; Istanbul, Turkey. Istanbul, Turkey: WPSA – Turkish Branch; 2004. p. 84
9. Tagirov M, Artemenko A, Tereshchenko A. Preservation of the poultry gene pool by cryoconservation. Agrarnoe reshenie [Agrarian Solution]; 2010. 10.
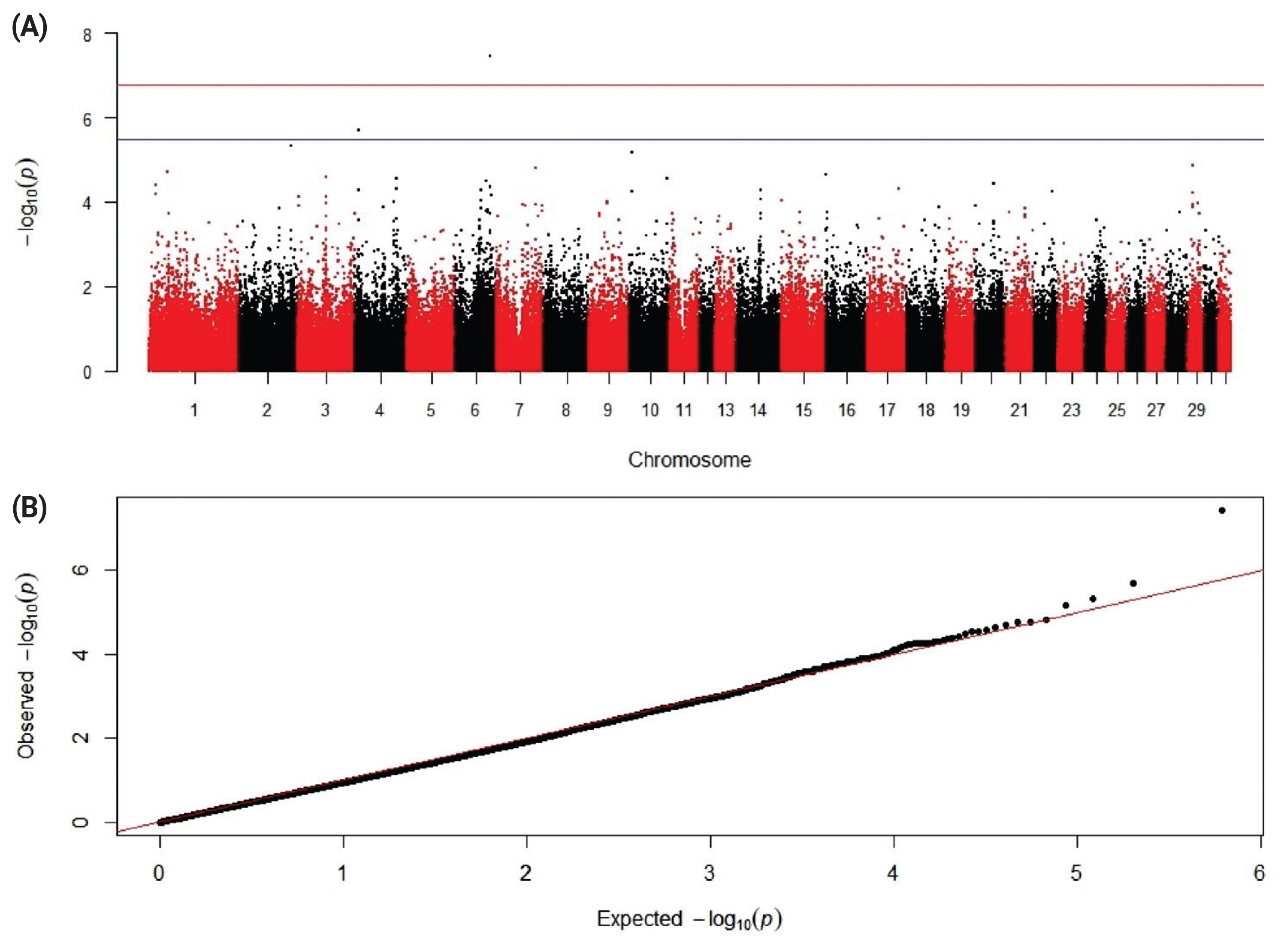
12. Kudinov AA, Dementieva N, Nikitkina E, Atroshchenko M, Musidrai A. 448 Late-breaking: GWAS analysis show QTL in horses which are characterized by sperm resistance to freezing. J Anim Sci 2019; 97:Suppl 3119–20.
https://doi.org/10.1093/jas/skz258.247

13. Greaves IK, Rens W, Ferguson-Smith MA, Griffin D, Marshall Graves JA. Conservation of chromosome arrangement and position of the X in mammalian sperm suggests functional significance. Chromosome Res 2003; 11:503–12.
https://doi.org/10.1023/a:1024982929452


14. Foster HA, Abeydeera LR, Griffin DK, Bridger JM. Non-random chromosome positioning in mammalian sperm nuclei, with migration of the sex chromosomes during late spermatogenesis. J Cell Sci 2005; 118:1811–20.
https://doi.org/10.1242/jcs.02301


17. Schrimpf R, Metzger J, Martinsson G, Sieme H, Distl O. Implication of FKBP6 for male fertility in horses. Reprod Domest Anim 2015; 50:195–9.
https://doi.org/10.1111/rda.12467


19. Usuga A, Rojano BA, Restrepo G. Association of the cysteine-rich secretory protein-3 (CRISP-3) and some of its polymorphisms with the quality of cryopreserved stallion semen. Reprod Fertil Dev 2018; 30:563–9.
https://doi.org/10.1071/RD17044


20. Restrepo G, Rojano B, Usuga A. Relationship of cysteine-rich secretory protein-3 gene and protein with semen quality in stallions. Reprod Domest Anim 2019; 54:39–45.
https://doi.org/10.1111/rda.13309


24. Mortimer D, Mortimer ST. Computer-aided sperm analysis (CASA) of sperm motility and hyperactivation. Carrell D, Aston K, editorsSpermatogenesis Methods in molecular biology. Totowa, NJ, USA: Humana Press; 2013. 927:p. 77–87.
https://doi.org/10.1007/978-1-62703-038-0_8

25. Suliman Y, Becker F, Wimmers K. Implication of transcriptome profiling of spermatozoa for stallion fertility. Reprod Fertil Dev 2018; 30:1087–98.
https://doi.org/10.1071/RD17188


26. Atroshchenko MM, Kalaschnikov VV, Bragina YY, Zaitsev AM. Comparative study of the structural integrity of spermatozoa in epididymal, ejaculated and cryopreserved semen of stallions. Sel’skokhozyaistvennaya Biol [Agric Biol] 2017; 52:274–81.
https://doi.org/10.15389/agrobiology.2017.2.274eng

27. Atroshchenko MM, Bragina EE, Zaitsev AM, et al. Conservation of genetic resources in horse breeding and major structural damages of sperm during semen cryopreservation in stallions. Nat Conserv Res 2019; 4:Suppl 278–82.
https://doi.org/10.24189/ncr.2019.024

30. R Core Team. R: A language and environment for statistical computing [Internet]. Vienna, Austria: R Foundation for Statistical Computing; 2021. [cited 2022 Jan 23]. Available from:
https://www.R-project.org/
31. Wickham H. ggplot2: elegant graphics for data analysis. New York, NY, USA: Springer-Verlag; 2016.
32. Zenkova D, Kamenev V, Sablina R, Artyomov M, Sergushichev A. Phantasus: Visual and interactive gene expression analysis [Internet]. Bioconductor. 2018. [cited 2022 Jan 23]. Available from:
https://doi.org/10.18129/B9.bioc.phantasus

40. Miranda-Vizuete A, Tsang KYY, Jiménez A, et al. Cloning and developmental analysis of murid spermatid-specific thioredoxin-2 (SPTRX-2), a novel sperm fibrous sheath protein and autoantigen. J Biol Chem 2003; 278:44874–85.
https://doi.org/10.1074/jbc.M305475200


43. Huet G, Rajakylä EK, Viita T, et al. Actin-regulated feedback loop based on Phactr4, PP1 and cofilin maintains the actin monomer pool. J Cell Sci 2013; 126:497–507.
https://doi.org/10.1242/jcs.113241


44. Fardilha M, Esteves SL, Korrodi-Gregório L, Pelech S, da Cruz E, Silva OA, da Cruz E, Silva E. Protein phosphatase 1 complexes modulate sperm motility and present novel targets for male infertility. Mol Hum Reprod 2011; 17:466–77.
https://doi.org/10.1093/molehr/gar004













 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Supplement
Supplement Print
Print




