Relationship between Single Nucleotide Polymorphisms in the Peroxisome Proliferator-Activated Receptor Gamma Gene and Fatty Acid Composition in Korean Native Cattle
Article information
Abstract
The peroxisome proliferator-activated receptor gamma (PPARγ) gene plays an important role in the biosynthesis process controlled by a number of fatty acid transcription factors. This study investigates the relationships between 130 single-nucleotide polymorphisms (SNPs) in the PPARγ gene and the fatty acid composition of muscle fat in the commercial population of Korean native cattle. We identified 38 SNPs and verified relationships between 3 SNPs (g.1159-71208 A>G, g.42555-29812 G>A, and g.72362 G>T) and the fatty acid composition of commercial Korean native cattle (n = 513). Cattle with the AA genotype of g.1159-71208 A>G and the GG genotype of g.42555-29812 G>A and g.72362 G>T had higher levels of monounsaturated fatty acids and carcass traits (p<0.05). The results revealed that the 3 identified SNPs in the PPARγ gene affected fatty acid composition and carcass traits, suggesting that these 3 SNPs may improve the flavor and quality of beef in commercial Korean native cattle.
INTRODUCTION
The fatty acid composition of livestock adipose tissue is recognized as an important carcass trait affecting meat quality. The quality of fat is determined by its fatty acid composition, and beef quality depends not only on the marbling score but also on fatty acid composition. Although beef marbling is an important factor, fatty acid composition also plays an important role in the eating quality of meat (Laborde et al., 2001). About 90% of the fatty acid composition of intramuscular fat from the longissimus muscle of beef cattle is accounted for by palmitic acid (16:0) and stearic acid (18:0) from saturated fatty acid (SFA) and oleic acid (18:1) from monounsaturated fatty acid (MUFA). Oleic acid accounts for the highest proportion of MUFAs in beef cattle and is positively correlated with marbling, beef flavor, tenderness, and soft fat (Melton et al., 1982; Kim et al., 2002; Lee et al., 2004; Lee et al., 2008; Smith et al., 2009). These factors are of considerable interest because of their implications for human health. Stearic acid (18:0) is a primary determinant of fat hardness (Smith et al., 1998; Wood et al., 2004; Chung et al., 2006).
Therefore, genetic factors that enhance the conversion of stearic acid into oleic acid are expected to increase tenderness, flavor, and marbling (Yang et al., 1999). The fatty acid biosynthesis process is controlled by a number of transcription factors such as peroxisome proliferator-activated receptor gamma (PPARγ), CCAAT/enhancer binding protein alpha (C/EBPA), and sterol regulatory element binding proteins (SREBPs). Many studies have reported that genetic factors in transcription factor genes are associated with fatty acid composition and the marbling score in Korean native cattle and Japanese Black cattle (Shin et al., 2007; Hoashi et al., 2007; Lee et al., 2008; Oh et al., 2012). These findings suggest that fatty acid composition may be controlled by genetic factors such as genes related to lipid synthesis and fatty acid metabolism (Narukami et al., 2011).
PPARγ, a member of the nuclear hormone receptor superfamily, acts as a ligand-inducible transcription factor (Spiegelman, 1998; Kersten et al., 2000). PPARγ forms a heterodimer complex with one of the three retinoid X receptor (RXR) proteins and then binds to PPAR-responsive elements (PPRE) in promoters of PPARγ target genes (Mangelsdorf and Evans, 1995; Schulman et al., 1998).
PPARs contain general structural features of other nuclear receptor family members, including a central DNA-binding domain, a C-terminal domain mediating ligand binding, and two transcription activation function motifs (Rosen and Spiegelman, 2001). PPARγ is expressed abundantly in adipocytes and plays a key role in mediating adipocyte differentiation and insulin sensitivity. Many adipocyte-specific genes such as adiponectin and perilipin contain response elements for PPARγ in their promoter regions (Iwaki et al., 2003; Nagai et al., 2004; Guan et al., 2005). In the absence of any ligand, nuclear corepressors bind PPAR/RXR heterodimers and recruit histone deactylases to down regulate gene transcription (Yu et al., 2005). The present study evaluates the relationships between single nucleotide polymorphisms (SNPs) of intron and exon regions in the PPARγ gene and fatty acid composition in Korean native cattle.
MATERIALS AND METHODS
Animals and phenotypic Data
A total of 513 commercial Korean native cattle samples from Gyeungsangbuk, 96, Korea, were included in the study where 10 sires were used to produce this half sib population. Feeding conditions (slaughter age, the fattening period, forage intake, and concentration) were controlled under livestock management of each region (n = 14). They were all steers slaughtered at 941±72 days of age, and the marbling score was measured 24 h after slaughter. First, the carcass was dissected at the last rib and the first lumber vertebra according to the Animal Product Grading System of Korea. Marbling scores were measured or scored in the left carcass cut across the vertebra between the last thoracic vertebra and the first lumbar vertebra. The marbling degree was scored from 1 to 9 with a mean of 5.43 such that the higher score, the more abundant the intramuscular fat (Table 1). Genomic DNA was extracted from the longissimus muscle by using the LaboPass Tissue Mini Kit (Cosmo Genetech, Seoul, Korea).
Fatty acid composition
Total lipids were extracted from approximately 500 mg of the longissimus muscle with chloroform/methanol (2/1, v/v) (Folch et al., 1957) and then methylated with sodium methylate (O’Keefe et al., 1968). Samples were filtered through a filter paper in a water bath (40°C). The filtrate was mixed with distilled water, from which a layer of methanol and water was removed. After the removal of chloroform and lipid layers by using nitrogen gas, the sample was treated with BF3-methanol (14%) and then subjected to transmethylation at 65°C. Fatty acid content was analyzed using gas-chromatography (PerkinElmer, Inc., Waltham, MA, USA). Fatty acids included myristic acid, palmitic acid, stearic acid, myristoleic acid, palmitoleic acid, oleic acid, linoleic acid, and linolenic acid. The composition and function of fatty acids were used as phenotypes in the analysis of the genetic relationship (Table 1).
Genotyping
130 SNPs in PPARγ were selected based on the reference SNP of the bovine PPARγ gene (GenBank No. NC_007320.5) in dbSNP. Genotypes of 10 SNPs were subjected to a preliminary analysis. Primers for the amplification and extension were designed for the single-base extension (SBE) (Vreeland et al., 2002) by using forward, reverse, and extension primer sequences (Supplementary Table 1). Reactions of the primer extension were performed using the SNaPshot ddNTP Primer Extension Kit (Applied Biosystems, Foster City, CA, USA). To purify reactions of the primer extension, mixtures containing exonuclease I and shrimp alkaline phosphatase were added to the reaction mixture. Samples were cultured at 37°C for 1 h and then inactivated at 72°C for 15 min. The polymerase chain reaction product was mixed with Genescan 120 LIZ standard and HiDi formamide (Applied Biosystems, USA), followed by denaturation at 95°C for 5 min. Electrophoresis was performed using the ABI PRISM 3130XL Genetic Analyzer, and then electrophoresis products were assayed using GeneMapper v.4.0 (Applied Biosystems, USA).
Statistic analysis
The significance of SNP allele frequency between the two groups (high and low) was analyzed using chi-square test for extremely varied 44 samples. Those SNP alleles for which there were significant differences in frequency were included in a linear discriminant analysis (Venables and Ripley, 2002) to develop a function that classified animals into two groups. Only influential fragments for discrimination (F>4.0) were included in the function.
The Hardy–Weinberg equilibrium (HWE) was tested for each SNP in the Korean native cattle population by using the chi-square statistic. Relationships between fatty acid composition and the marbling score for individual SNPs were analyzed using the following mixed model based on SPSS v19.0 (SPSS Inc., Chicago, IL, USA): Yijkl = μ+Si+Pj+Gk+b_age+eijk, where Yijkl is the phenotype, μ is the overall mean, Si is the random effect of sire i with the assumption of an independent and identical normal distribution, Pj is the fixed effect of calving place j (14 classes), Gk is the fixed effect of genotype k, b is the regression coefficient, age is a covariate for age in days at slaughter, and eijk is a random residual assumed to have an independent and identical normal distribution. Significant differences in mean values between different genotypes were calculated using Duncan’s multiple-range test. Here p<0.05 was considered to be significant. Additive and dominant effects were estimated using the REG procedure in SAS version 9.2. For the haplotype analysis, the linkage disequilibrium (LD) between SNP pairs was measured according to D′ and r2 based on the HaploView software package (MIT/Harvard Broad Institute, Cambridge, MA, USA).
RESULTS AND DISCUSSION
The SNP selection of the PPARγ gene and the haplotype analysis of Hanwoo
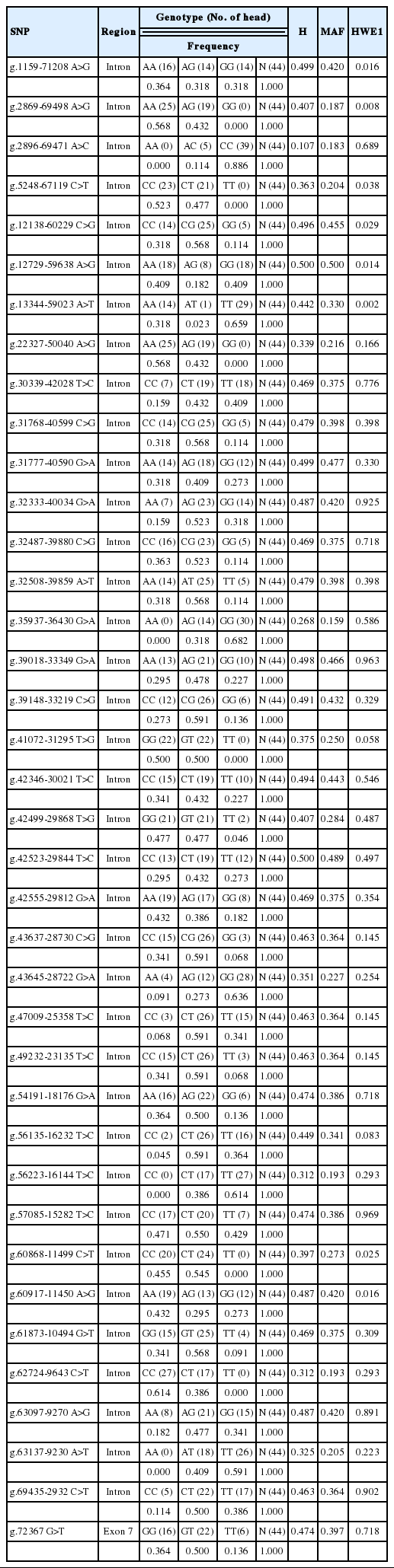
All 130 SNPs at the intron and exon were identified by an NCBI-based analysis to evaluate the relationships between SNPs in the PPARγ gene and fatty acid composition (Figure 1). Here 129 SNPs were located at the intron, and 1, at exon 7. In addition, the phenotypic mean and standard deviation, the min and max of carcass traits, fatty acid composition, and the fatty acid index were determined for cattle (Table 1). By the SBE for the verification of 130 SNPs in 44 cattle for the high- and low-MUFA groups of Korean native cattle, 38 SNPs were identified: 37 SNPs at the intron and 1 at exon 7. Statistical analyses were conducted to determine genotypic and allelic frequency, heterozygotes, minor-allele frequency (MAF), and the HWE. MAF was lower than 0.100, and segregation was not clearly observed following the LD analysis, which reduced analytical accuracy (Eberle et al., 2006; Lee et al., 2006). The 38 identified SNPs were found to have MAF values greater than 0.100, and their genotypes did not deviate from the HWE (p>0.05; Table 2) (Falconer and Mackay, 1996).

130 SNPs and linkage disequilibrium (LD) values for 3 SNPs in the bovine PPARγ gene. The HaploView plot is color-coded using the following scheme: White represents r2 = 0, and red signifies r2 = 1. Numbers in cells are R2 values. However, R2 values of 1.0 are not shown. SNPs, single nucleotide polymorphisms; PPARγ, peroxisome proliferator-activated receptor gamma.
Discrimination efficiency
Table 3 shows the efficiency of discrimination for the 38 SNPs in the high- and low-MUFA groups of Korean native cattle (n = 44). To classify 44 extreme animals into the high and low MUFAs groups, discriminant coefficients were calculated for the 38 SNPs, and these coefficients were derived using information on the SNP genotype and the high- or low-MUFA group. Among these SNPs, the g.1159-71208 A>G, g.42555-29812 G>A, and g.72367 G>T SNPs had the highest discriminant coefficients (−1.094, 0.700, and −0.941, respectively). Three SNPs showed significant relationships with MUFAs (p<0.05, F>4.0). This is equivalent to a discriminative error of 2.1% and a high correlation ratio of 0.91. These results indicated that the discriminant function successfully separated extreme Korean native cattle (Tsuji et al., 2004). Therefore, the g.1159-71208 A>G, g.42555-29812 G>A, and g.72367 G>T SNPs were selected for large-scale genotyping in commercial Korean native cattle (n = 513). Based on these results, a pairwise LD analysis of these 3 SNPs showed that the PPARγ gene cannot be conducted block (Figure 1).
Relationships with fatty acid composition
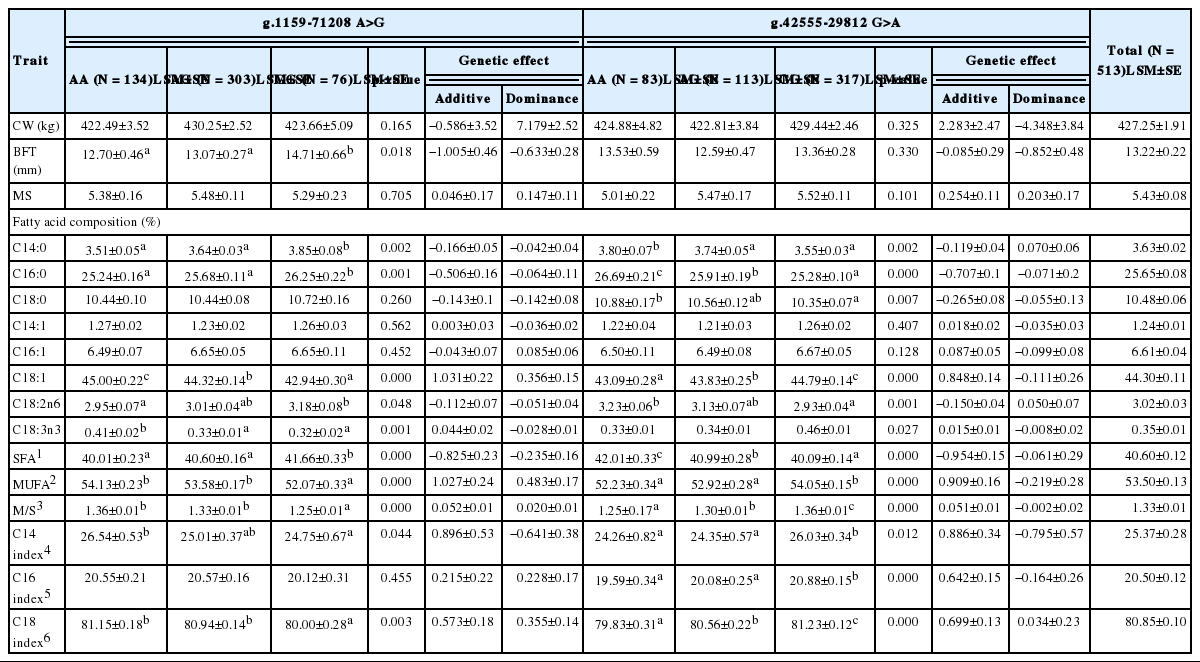
Previous studies have shown that the content of unsaturated fatty acid (UFA) (Alexander et al., 2007) and a high MUFA level have significant effects on beef flavor (Hoashi et al., 2007). Based on these reports, fatty acid composition, carcass phenotypes, and genetic effects were examined for a single Korean native cattle genotype (Tables 4 and 5). The results revealed significant differences according to the SNP of the PPARγ gene.
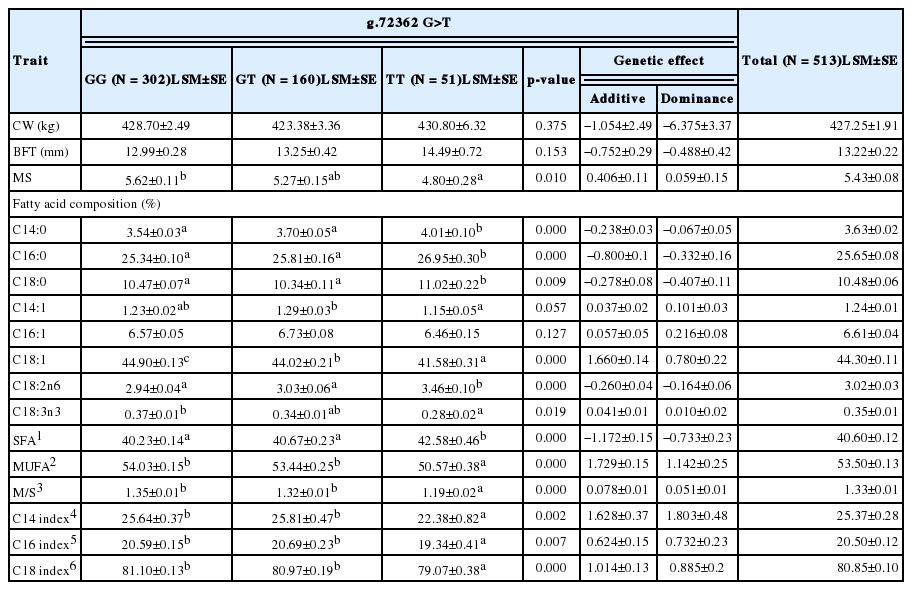
According to the results for carcass traits and two SNPs (g.1159-71208 A>G and g.72362 G>T), g.1159-71208 A>G were significantly related to backfat thickness (BFT), and g.72362 G>T to the marbling score (MS). The relationships between carcass traits and each SNP were based on the p-value (p<0.05). Yue et al. (2011) and Shin et al. (2006) reported significant effects of the bovine PPARγ genotype (p<0.05) on tenderness, BFT and water-holding capacity in Chinese indigenous bovine and on the longissimus muscle area and carcass weight in Korean native cattle. And All the mRNA region of the PPARγ gene within 760 individuals of four Chinese cattle breeds was scanned and four SNPs (−110G>C, −27C>T and +20A>G) of the PPARγ were detected in three Chinese indigenous cattle breeds (Qinchuan, Nangyang and Jiaxian cattle), rather than Chinese Holstein cattle. The mutations −110G>C, −27C>T and +20A>G located in the Exon1 of the PPARG and were under LD. The individuals with these three mutations had smaller body measurements (Hua et al., 2011). In the case of these five SNPs, they were located at intron 3 (C/T, 292), intron 5 (C/T, 1,064) and exon 1 (−110G>C, −27C>T, +20A>G) in the PPARγ gene. Although the position of these SNPs and the breed of cattle in these studies are different from those in the present study, their findings are consistent with this study’s results.
As shown in Table 4 and 5, SFA in the g.1159-71208 A>G, g.42555-29812 G>A, and g.72362 G>T SNPs was observed at 41.66%, 42.01%, and 42.58% in genotypes of GG, AA, and TT, respectively (p<0.05). By contrast, oleic acid (C18:1) levels were significantly related to the AA genotype of g.1159-71208 A>G, the GG genotype of g.42555-29812 G>A, and the GG genotype of g.72362 G>T (p<0.01). In addition, the AA and GG genotypes of g.1159-71208 A>G and g.42555-29812 G>A and the GG genotype of g.72362 G>T were related to the highest oleic acid content. MUFAs account for a high proportion of fatty acid composition and are an important factor influencing meat quality. Among MUFAs, oleic acid (C18:1) is one of the major MUFAs in beef cattle and the aroma source of cooked beef (Melton et al., 1982; Mandell et al., 1998). MUFA levels (54.13%, 54.05%, and 54.03%, respectively) showed the same trends as those for oleic acid (p<0.05). Clinical studies have shown that long-chain omega-3 polyunsaturated fatty acids (n-3 PUFA; linolenic acid) have beneficial effects on human health. On the other hand, high levels of omega-6 PUFAs (n-6 PUFAs; linoleic acid) are closely related to certain human diseases such as cancer, cardiovascular disease, and various mental disorders (Reidiger et al., 2009). In the present study, levels of PUFAs, which included linoleic acid (C18:2n6; omega-6) and linolenic acid (C18:3n3; omega-3), were related to the three SNPs evaluated (p<0.05). Linolenic acid had the highest content in GG, AA, and TT genotypes, but the opposite trend was observed for linoleic acid. Variations in linolenic acid showed trends similar to those in oleic acid and MUFA content. However, the opposite trend was observed for linoleic acid. These results suggest that the consumption of meat from Korean native cattle with specific SNPs that alter levels of different fatty acids may enhance human health.
As shown in Table 4, g.1159-71208 A>G and g.42555-29812 G>A had additive effects on oleic acid, linoleic acid, linolenic acid, SFAs, and MUFAs (p<0.05). In addition, g.72362 G>T had an additive effect on MS, oleic acid, SFAs, and MUFAs. In particular, oleic acid (C18:1), an important factor influencing beef flavor, had significant additive effects (1.031, 0.848, and 1.660, respectively) on quantitative traits, and these effects were positive.
In general, PPARγ is a type of nuclear receptor superfamily with the N-Terminal domain, the DNA-binding domain, and the ligand-binding domain. Previous studies have reported that PPARγ, whose ligand-binding domain is bound by the ligand, forms a dimer with RXR, which is a 9-cis-retinoic acid receptor, and thus that it controls adipocyte differentiation by combining with the peroxisome proliferator response element (PPRE), which belongs to the area of the cis-acting element relevant to adipocyte differentiation (Kodera et al., 2000; Christine et al., 2005; Shanika et al., 2009). This ligand-binding domain consists of 154 amino acids ranging from the 321st valine to the 474th valine in Hanwoo (Jeoung et al., 2004). In particular, g.72367 G>T SNP (Gln1344His) is the 448th glutamine included in the ligand-binding domain. In addition, it may become a non-synonymous SNP that changes into histidine from glutamine as the CAG nucleotide changes into CAT (Oh et al., 2012). This suggests that fatty acid content may vary depending on the level of change in the amino acid of the ligand-binding domain because it affects adipocyte differentiation.
In conclusion, Korean native cattle with AA, GG, and GG genotypes at the g.1159-71208 A>G, g.42555-29812 G>A, and g.72362 G>T SNPs in the PPARγ gene had higher proportions of MUFAs and lower proportions of SFAs. In addition, animals with the GG genotype of g.72362 G>T had higher MSs. These individual SNPs may be effective genetic targets for improving beef quality and have considerable influence on the Korean native cattle industry.
Supplementary Data
ACKNOWLEDGMENTS
This research was supported by the Yeungnam University Research Grant in 2014.
Notes
CONFLICT OF INTEREST
We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.