 |
 |
| Anim Biosci > Volume 35(5); 2022 > Article |
|
Abstract
Objective
Methods
Results
Notes
Figure┬Ā1
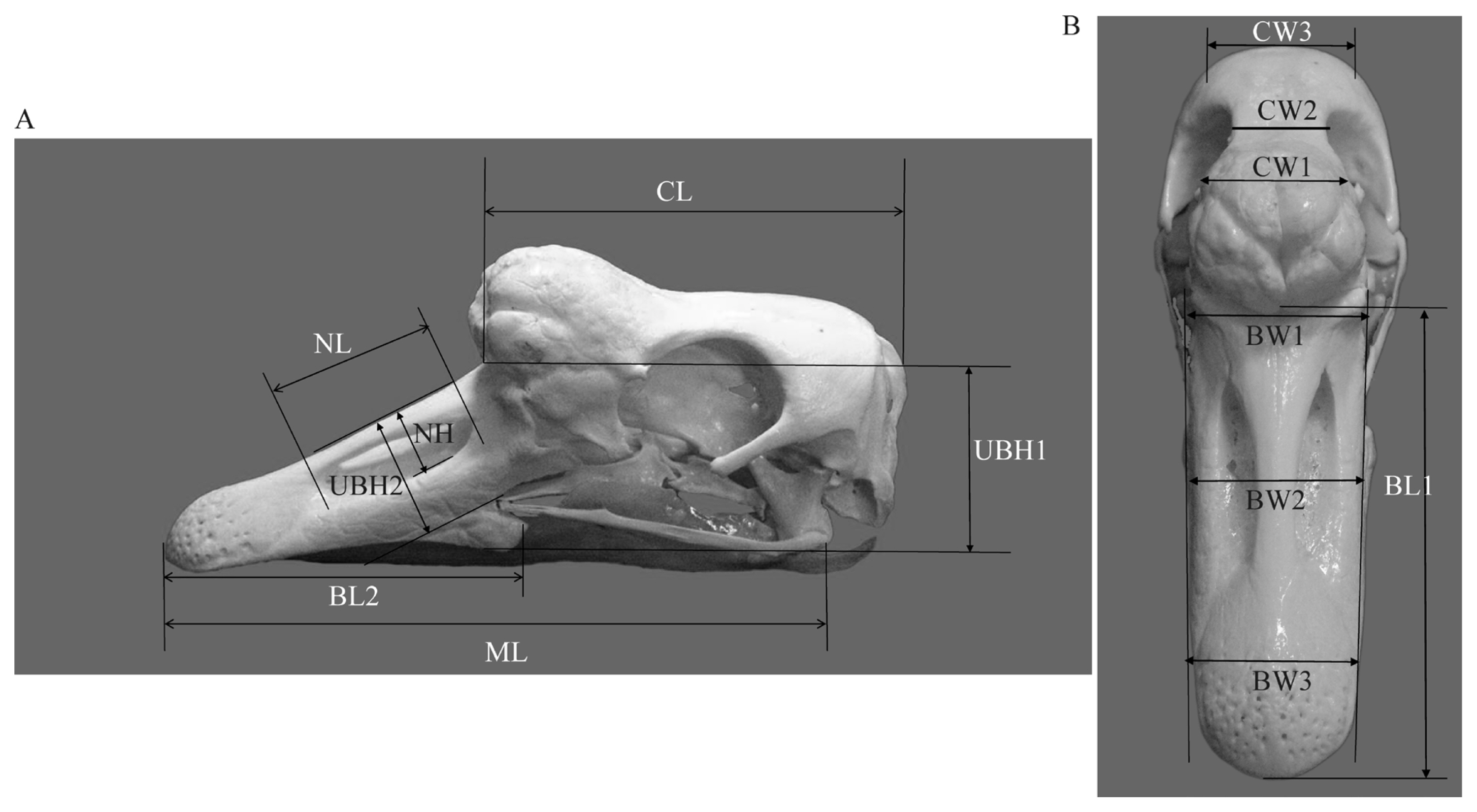
Figure┬Ā2

Figure┬Ā3
Table┬Ā1
| Knob types | BL1 | BL2 | BW1 | BW2 | BW3 | CW1 | CW2 | CW3 | ML | CL | UBH1 | UBH2 | NL | NH |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Knob geese (138) | 6.62┬▒0.51* | 7.19┬▒0.57 | 3.18┬▒0.21* | 2.74┬▒0.17* | 2.51┬▒0.15* | 2.89┬▒0.26* | 2.29┬▒0.22* | 3.08┬▒0.24* | 13.80┬▒0.91* | 8.16┬▒0.45* | 3.13┬▒0.27* | 1.88┬▒0.25* | 2.91┬▒0.24* | 0.81┬▒0.15 |
| Non-knob geese (166) | 6.29┬▒0.39 | 6.89┬▒0.46 | 3.01┬▒0.18 | 2.61┬▒0.14 | 2.41┬▒0.10 | 2.73┬▒0.18 | 2.16┬▒0.19 | 2.94┬▒0.25 | 13.25┬▒0.66 | 7.91┬▒0.36 | 3.01┬▒0.24 | 1.78┬▒0.19 | 2.83┬▒0.22 | 0.80┬▒0.14 |
The number behind the knob types represented the number of individuals. Centimeter (cm) was used as the unit for length, height and width.
BL1, beak length 1; BL2, beak length 2; BW1, beak width 1; BW2, beak width 2; BW3, beak width 3; CW1, cranial width 1; CW2, cranial width 2; CW3, cranial width 3; ML, maxillary length; CL, cranial length; UBH1, height of upper beak 1; UBH2, height of upper beak 2; NL, nostril length; NH, nostril height.
Table┬Ā2
The number behind the genotype represented the number of individuals. Centimeter (cm) was used as the unit for length, height and width.
BL1, beak length 1; BL2, beak length 2; BW1, beak width 1; BW2, beak width 2; BW3, beak width 3; CW1, cranial width 1; CW2, cranial width 2; CW3, cranial width 3; ML, maxillary length; CL, cranial length; UBH1, height of upper beak 1; UBH2, height of upper beak 2; NL, nostril length; NH, nostril height.
Table┬Ā3
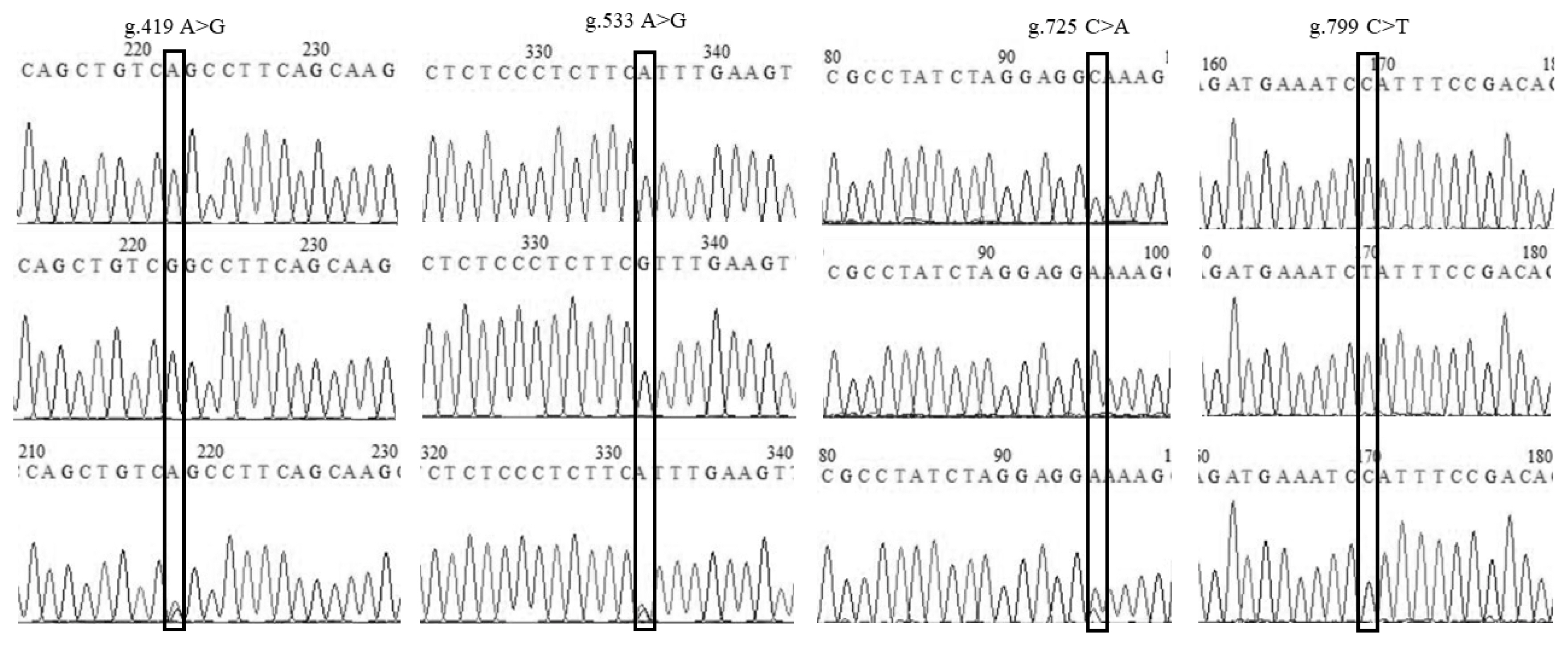
| Loci | Genotypes | BL1 | BL2 | BW1 | BW2 | BW3 | CW1 | CW2 | CW3 | ML | CL | UBH1 | UBH2 | NL | NH |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| g.419 G>A | GG (56) | 6.32┬▒0.39 | 6.92┬▒0.47 | 3.00┬▒0.18 | 2.60┬▒0.15 | 2.40┬▒0.10 | 2.70┬▒0.21 | 2.15┬▒0.19 | 2.96┬▒0.23 | 13.31┬▒0.67 | 7.91┬▒0.39 | 3.02┬▒0.24 | 1.77┬▒0.18 | 2.82┬▒0.21 | 0.80┬▒0.13 |
| AA (40) | 6.33┬▒0.38 | 6.92┬▒0.39 | 3.09┬▒0.19* | 2.66┬▒0.15* | 2.43┬▒0.12 | 2.79┬▒0.16 | 2.19┬▒0.19 | 2.91┬▒0.31 | 13.39┬▒0.69* | 7.99┬▒0.32 | 3.03┬▒0.26 | 1.75┬▒0.17 | 2.92┬▒0.19* | 0.76┬▒0.10 | |
| GA (70) | 6.25┬▒0.39 | 6.86┬▒0.49 | 2.98┬▒0.16 | 2.59┬▒0.13 | 2.40┬▒0.10 | 2.72┬▒0.17 | 2.16┬▒0.20 | 2.95┬▒0.23 | 13.13┬▒0.61 | 7.86┬▒0.35 | 2.98┬▒0.24 | 1.80┬▒0.21 | 2.79┬▒0.23 | 0.82┬▒0.16* | |
| g.533 G>A | GG (56) | 6.32┬▒0.39 | 6.92┬▒0.47 | 3.00┬▒0.18 | 2.60┬▒0.15 | 2.40┬▒0.10 | 2.70┬▒0.21 | 2.15┬▒0.19 | 2.96┬▒0.23 | 13.31┬▒0.67 | 7.91┬▒0.39 | 3.02┬▒0.24 | 1.77┬▒0.18 | 2.82┬▒0.21 | 0.80┬▒0.13 |
| AA (40) | 6.33┬▒0.38 | 6.92┬▒0.39 | 3.09┬▒0.19* | 2.66┬▒0.15* | 2.43┬▒0.12 | 2.79┬▒0.16 | 2.19┬▒0.19 | 2.91┬▒0.31 | 13.39┬▒0.69* | 7.99┬▒0.32 | 3.03┬▒0.26 | 1.75┬▒0.17 | 2.92┬▒0.19* | 0.76┬▒0.10 | |
| GA (70) | 6.25┬▒0.39 | 6.86┬▒0.49 | 2.98┬▒0.16 | 2.59┬▒0.13 | 2.40┬▒0.10 | 2.72┬▒0.17 | 2.16┬▒0.20 | 2.95┬▒0.23 | 13.13┬▒0.61 | 7.86┬▒0.35 | 2.98┬▒0.24 | 1.80┬▒0.21 | 2.79┬▒0.23 | 0.82┬▒0.16* | |
| g. 725 A>C | AA (56) | 6.32┬▒0.39 | 6.91┬▒0.47 | 2.99┬▒0.18 | 2.61┬▒0.14 | 2.41┬▒0.11 | 2.70┬▒0.21 | 2.16┬▒0.19 | 2.97┬▒0.23 | 13.25┬▒0.63 | 7.94┬▒0.36 | 3.02┬▒0.25 | 1.78┬▒0.17 | 2.81┬▒0.21 | 0.81┬▒0.13 |
| CC (39) | 6.32┬▒0.38 | 6.91┬▒0.39 | 3.09┬▒0.19* | 2.65┬▒0.15* | 2.43┬▒0.11 | 2.78┬▒0.16* | 2.19┬▒0.18 | 2.91┬▒0.31 | 13.36┬▒0.68 | 7.99┬▒0.32 | 3.04┬▒0.26 | 1.75┬▒0.17 | 2.92┬▒0.19* | 0.76┬▒0.10 | |
| AC (71) | 6.25┬▒0.39 | 6.87┬▒0.48 | 2.99┬▒0.15 | 2.59┬▒0.13 | 2.40┬▒0.10 | 2.72┬▒0.17 | 2.15┬▒0.20 | 2.94┬▒0.23 | 13.19┬▒0.66 | 7.85┬▒0.37 | 2.98┬▒0.23 | 1.79┬▒0.21 | 2.80┬▒0.23 | 0.81┬▒0.16 | |
| g.799 T>C | TT (55) | 6.33┬▒0.38 | 6.94┬▒0.46 | 3.01┬▒0.18 | 2.62┬▒0.14 | 2.41┬▒0.11 | 2.71┬▒0.21 | 2.16┬▒0.19 | 2.96┬▒0.23 | 13.32┬▒0.66 | 7.92┬▒0.39 | 3.02┬▒0.24 | 1.78┬▒0.18 | 2.82┬▒0.21 | 0.81┬▒0.13 |
| CC (39) | 6.32┬▒0.38 | 6.91┬▒0.39 | 3.09┬▒0.19* | 2.65┬▒0.15* | 2.43┬▒0.11 | 2.78┬▒0.16* | 2.19┬▒0.18 | 2.91┬▒0.31 | 13.36┬▒0.68 | 7.99┬▒0.32 | 3.04┬▒0.26 | 1.75┬▒0.17 | 2.92┬▒0.19* | 0.76┬▒0.10 | |
| TC (72) | 6.24┬▒0.40 | 6.84┬▒0.49 | 2.98┬▒0.16 | 2.58┬▒0.13 | 2.39┬▒0.10 | 2.72┬▒0.17 | 2.15┬▒0.20 | 2.95┬▒0.23 | 13.13┬▒0.63 | 7.86┬▒0.34 | 2.98┬▒0.24 | 1.79┬▒0.21 | 2.79┬▒0.23 | 0.81┬▒0.14 |
The number behind the genotype represented the number of individuals. Centimeter (cm) was used as the unit for length, height and width.
BL1, beak length 1; BL2, beak length 2; BW1, beak width 1; BW2, beak width 2; BW3, beak width 3; CW1, cranial width 1; CW2, cranial width 2; CW3, cranial width 3; ML, maxillary length; CL, cranial length; UBH1, height of upper beak 1; UBH2, height of upper beak 2; NL, nostril length; NH, nostril height.
Table┬Ā4
| Hap/Dip | BL1 | BL2 | BW1 | BW2 | BW3 | CW1 | CW2 | CW3 | ML | CL | UBH1 | UBH2 | NL | NH |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GGAT | 6.35┬▒0.39 | 6.94┬▒0.47 | 3.00┬▒0.18 | 2.61┬▒0.14 | 2.40┬▒0.10 | 2.70┬▒0.21 | 2.16┬▒0.19 | 2.97┬▒0.23 | 13.27┬▒0.64 | 7.93┬▒0.37 | 3.03┬▒0.24 | 1.78┬▒0.18 | 2.82┬▒0.22 | 0.81┬▒0.13* |
| AACC | 6.32┬▒0.38 | 6.91┬▒0.39 | 3.09┬▒0.19* | 2.65┬▒0.15 | 2.43┬▒0.11 | 2.78┬▒0.16* | 2.19┬▒0.18 | 2.91┬▒0.31 | 13.36┬▒0.68 | 7.99┬▒0.32 | 3.04┬▒0.26 | 1.75┬▒0.17 | 2.92┬▒0.19* | 0.76┬▒0.10 |
| AACC-AACC | 6.32┬▒0.38 | 6.91┬▒0.39 | 3.09┬▒0.19* | 2.65┬▒0.15* | 2.43┬▒0.11 | 2.78┬▒0.16* | 2.19┬▒0.18 | 2.91┬▒0.31 | 13.36┬▒0.68 | 7.99┬▒0.32 | 3.04┬▒0.26 | 1.75┬▒0.17 | 2.92┬▒0.19* | 0.76┬▒0.10 |
| GGAT-AACC | 6.26┬▒0.40 | 6.86┬▒0.49 | 2.99┬▒0.16 | 2.59┬▒0.12 | 2.39┬▒0.09 | 2.72┬▒0.17 | 2.15┬▒0.20 | 2.95┬▒0.23 | 13.12┬▒0.62 | 7.85┬▒0.36 | 2.99┬▒0.24 | 1.80┬▒0.22 | 2.79┬▒0.24 | 0.82┬▒0.17* |
| GGAT-GGAT | 6.35┬▒0.39 | 6.94┬▒0.47 | 3.00┬▒0.18 | 2.61┬▒0.14 | 2.40┬▒0.10 | 2.70┬▒0.21 | 2.16┬▒0.19 | 2.97┬▒0.23 | 13.27┬▒0.64 | 7.93┬▒0.37 | 3.03┬▒0.24 | 1.78┬▒0.18 | 2.82┬▒0.22 | 0.81┬▒0.13 |
BL1, beak length 1; BL2, beak length 2; BW1, beak width 1; BW2, beak width 2; BW3, beak width 3; CW1, cranial width 1; CW2, cranial width 2; CW3, cranial width 3; ML, maxillary length; CL, cranial length; UBH1, height of upper beak 1; UBH2, height of upper beak 2; NL, nostril length; NH, nostril height.






 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Supplement
Supplement Print
Print





