8. Lukaszewski MA, Eberlé D, Vieau D, Breton C. Nutritional manipulations in the perinatal period program adipose tissue in offspring. Am J Physiol Endocrinol Metab 2013; 305:e1195–207.
https://doi.org/10.1152/ajpendo.00231.2013


13. Pope M, Budge H, Symonds ME. The developmental transition of ovine adipose tissue through early life. Acta Physiol 2014; 210:20–30.
https://doi.org/10.1111/apha.12053

15. Satterfield MC, Dunlap KA, Bazer FW, Wu G. Arginine nutrition and fetal brown adipose tissue development in nutrient-restricted sheep. Amino Acids 2013; 45:489–99.
https://doi.org/10.1007/s00726-011-1168-8


16. Nissen PM, Oksbjerg N. Quantification of prenatal effects on productivity in pigs. Greenwood PL, Bell AW, Vercoe PE, editorsManaging the prenatal environment to enhance livestock productivity. New York, USA: Springer; 2010. p. 37–70.

17. Xu YM. Death causes and countermeasures of newborn lambs in northern cold season. Zhongguo Dongwu Baojian 2013; 3:56–7.
18. Budge H, Dandrea J, Mostyn A, et al. Differential effects of fetal number and maternal nutrition in late gestation on prolactin receptor abundance and adipose tissue development in the neonatal lamb. Pediatr Res 2003; 53:302–8.


19. Gao F, Liu YC, Zhang CZ, Zhang ZH, Song SS. Effect of intrauterine growth restriction during late pregnancy on the growth performance, blood components, immunity and anti-oxidation capability of ovine fetus. Livest Sci 2013; 155:435–41.
https://doi.org/10.1016/j.livsci.2013.04.016

20. Symonds ME, Budge H, Stephenson T, McMillen IC. Fetal endocrinology and development--manipulation and adaptation to long-term nutritional and environmental challenges. Reproduction 2001; 121:853–62.
https://doi.org/10.1530/rep.0.1210853


21. Symonds ME, Mostyn A, Pearce S, Budge H, Stephenson T. Endocrine and nutritional regulation of fetal adipose tissue development. J Endocrinol 2003; 179:293–9.
https://doi.org/10.1677/joe.0.1790293


23. Satterfield MC, Wu G. Brown adipose tissue growth and development: significance and nutritional regulation. Front Biosci (Landmark Ed) 2011; 16:1589–608.
https://doi.org/10.2741/3807


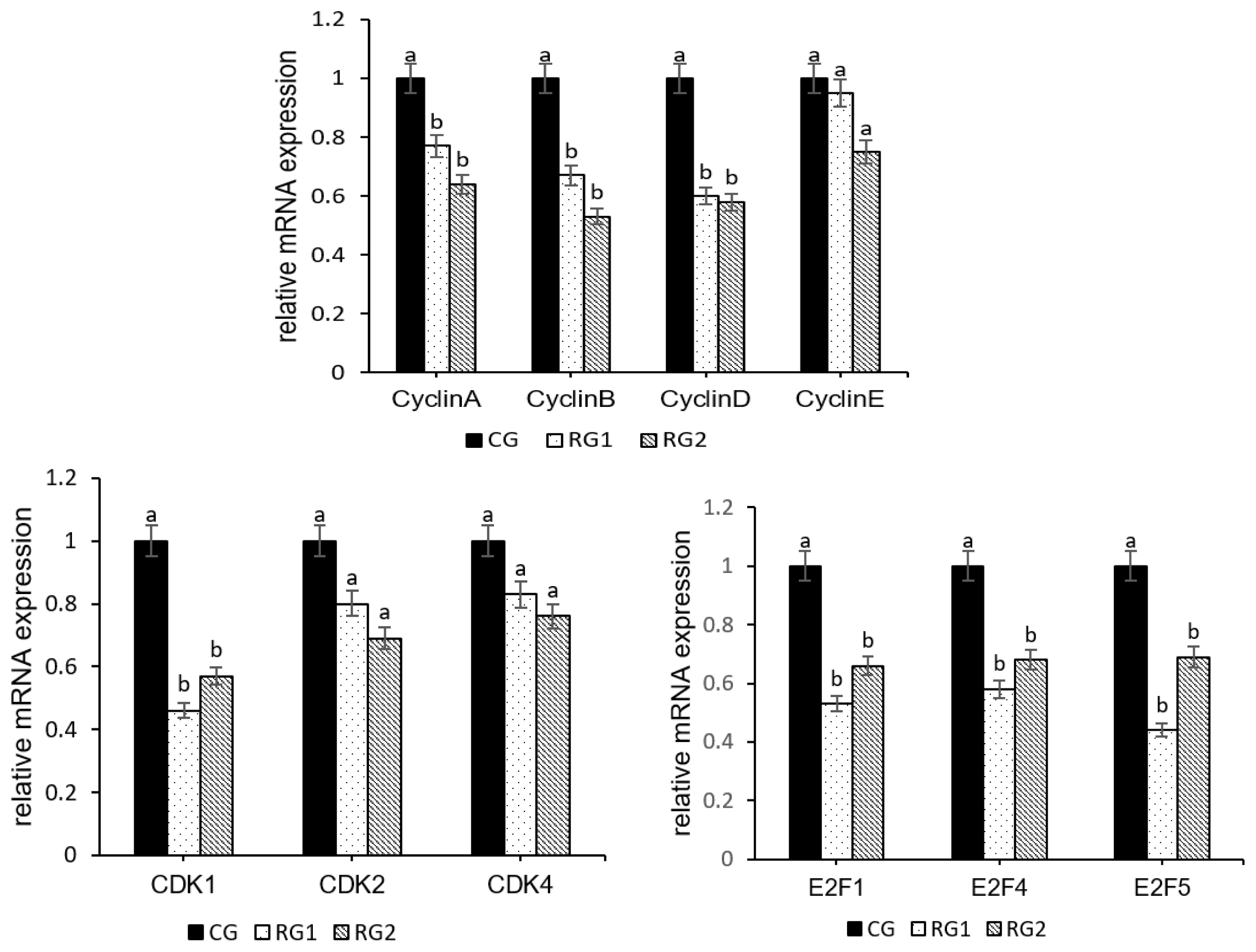
30. Calbo J, Parreno M, Sotillo E, et al. G1 Cyclin/cyclin-dependent kinase-coordinated phosphorylationof endogenous pocket proteins differentially regulates their interactions with E2F4 and E2F1 and gene expression. J Biol Chem 2002; 277:50263–74.
https://doi.org/10.1074/jbc.M209181200


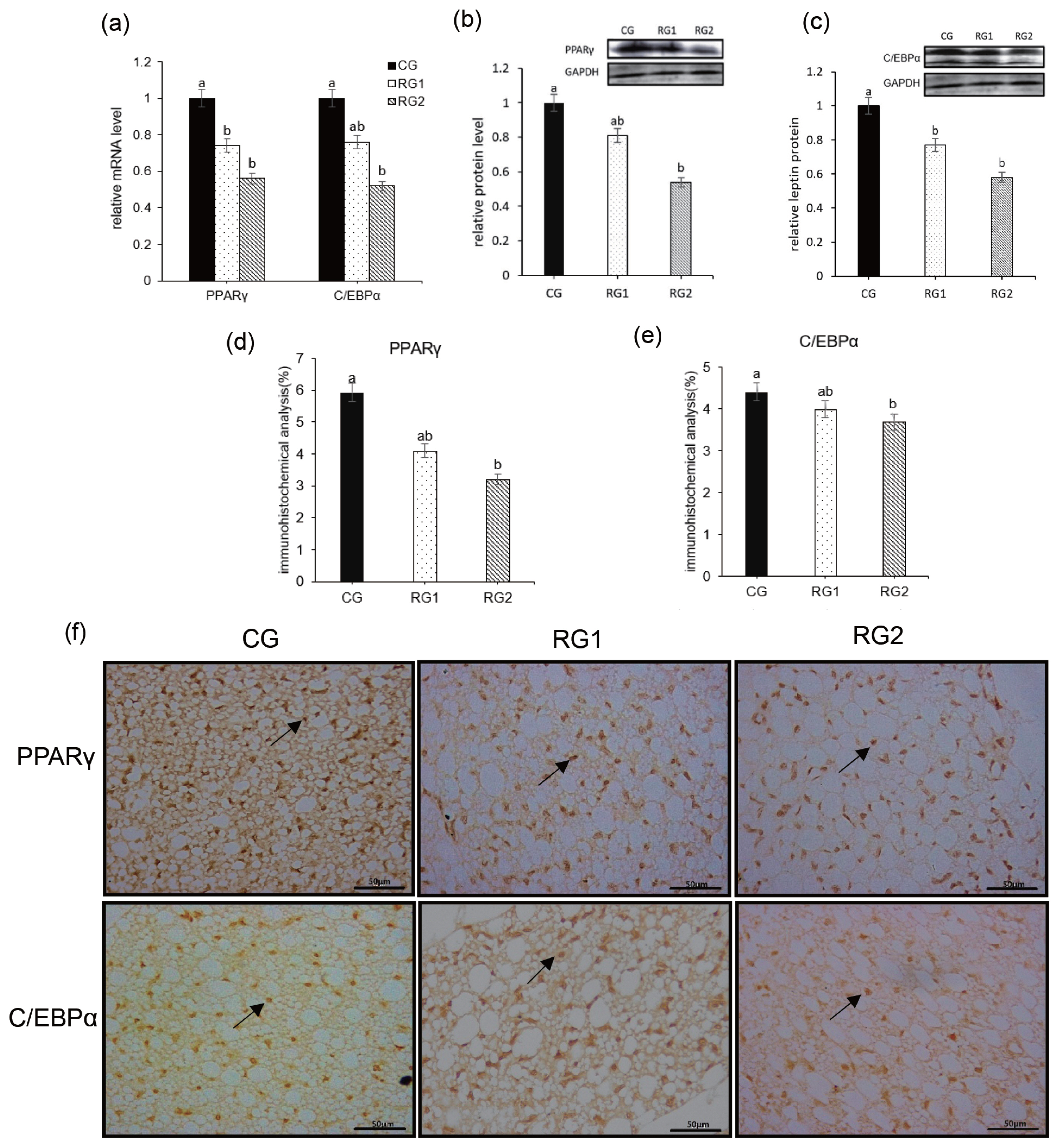
34. Duffield JA, Vuocolo T, Tellam R, et al. Intrauterine growth restriction and the sex specific programming of leptin and peroxisome proliferator-activated receptor gamma (PPARgamma) mRNA expression in visceral fat in the lamb. Pediatr Res 2009; 66:59–65.
https://doi.org/10.1203/PDR.0b013e3181a7c121


39. Elsen M, Raschke S, Tennagels N, et al. BMP4 and BMP7 induce the white-to-brown transition of primary human adipose stem cells. Am J Physiol Cell Physiol 2014; 306:C431–40.
https://doi.org/10.1152/ajpcell.00290.2013











 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print




