 |
 |
| Anim Biosci > Volume 28(10); 2015 > Article |
|
Abstract
Genetic parameters of Berkshire pigs for reproduction, carcass and meat quality traits were estimated using the records from a breeding farm in Korea. For reproduction traits, 2,457 records of the total number of piglets born (TNB) and the number of piglets born alive (NBA) from 781 sows and 53 sires were used. For two carcass traits which are carcass weight (CW) and backfat thickness (BF) and for 10 meat quality traits which are pH value after 45 minutes (pH45m), pH value after 24 hours (pH24h), lightness in meat color (LMC), redness in meat color (RMC), yellowness in meat color (YMC), moisture holding capacity (MHC), drip loss (DL), cooking loss (CL), fat content (FC), and shear force value (SH), 1,942 pig records were used to estimate genetic parameters. The genetic parameters for each trait were estimated using VCE program with animal model. Heritability estimates for reproduction traits TNB and NBA were 0.07 and 0.06, respectively, for carcass traits CW and BF were 0.37 and 0.57, respectively and for meat traits pH45m, pH24h, LMC, RMC, YMC, MHC, DL, CL, FC, and SH were 0.48, 0.15, 0.19, 0.36, 0.28, 0.21, 0.33, 0.45, 0.43, and 0.39, respectively. The estimate for genetic correlation coefficient between CW and BF was 0.27. The Genetic correlation between pH24h and meat color traits were in the range of ŌłÆ0.51 to ŌłÆ0.33 and between pH24h and DL and SH were ŌłÆ0.41 and ŌłÆ0.32, respectively. The estimates for genetic correlation coefficients between reproductive and meat quality traits were very low or zero. However, the estimates for genetic correlation coefficients between reproductive traits and drip and cooking loss were in the range of 0.12 to 0.17 and ŌłÆ0.14 to ŌłÆ0.12, respectively. As the estimated heritability of meat quality traits showed medium to high heritability, these traits may be applicable for the genetic improvement by continuous measurement. However, since some of the meat quality traits showed negative genetic correlations with carcass traits, an appropriate breeding scheme is required that carefully considers the complexity of genetic parameters and applicability of data.
Meat productivity traits such as carcass weight (CW) and backfat thickness and reproductive traits such as total litter size and the number of piglets born alive are very important economic traits in swine. Therefore, until now selection of pigs has been focused on these traits. However, due to the improved eating habits of consumers, breeding objectives have also put weight on meat quality (van Wijk et al., 2005). As customersŌĆÖ demands for pork depend upon the meat quality and its physical and biochemical components (Bonneau and Lebret, 2010), it is time that genetic improvement is required not only for productive and reproductive traits but also meat quality traits. Meat quality traits are highly heritable, causing an efficient selection response (Newcom et al., 2002). Therefore, meat quality is a trait applicable for the genetic improvement and should be one of the major selection benchmarks for breeding process in the swine industry (Luo et al., 2012).
It has been reported that there is a negative or very low genetic correlation between reproductive traits and meat productivity and quality traits (Ducos and Bidanel, 1996; Clutter and Brascamp, 1998; Chen et al. 2003b; Holm et al., 2004). When traits have a negative genetic correlation, it is difficult to select simultaneously for both traits. The establishment of the breeding improvement direction should be accurate when evaluating the genetic performance of sires and the most important preceding condition for the establishment of breeding improvement direction is the precise estimation of genetic parameters.
Recently, production of Berkshire pigs has increased in Korea because of their excellent meat qualities such as marbling, juiciness, tenderness and flavor. For the genetic improvement of Korean Berkshire pigs, selection should simultaneously consider productive, reproductive and meat quality traits, which requires the precise estimation of genetic parameters.
This experiment was conducted to estimate genetic correlations among major economic traits such as reproductive, carcass and meat quality traits in the Berkshire pig population and to establish selection and improvement direction thereafter.
A total of 2,511 records on the reproductive traits of Berkshire sows farrowed from 2007 to 2013 were collected from Dasan Pig Breeding Farm (Namwon, Korea). Data that lacked litter size, abortion during pregnancy and farrowing difficulty were excluded from the study. Therefore, for the final analysis only 2,457 records composed of the total number of piglets born (TNB) and the numbers of piglets born alive (NBA) from 781 sows were used, while the total number of sires were 53 heads. The mean and maximum for the parity of sows were 3.27 and 10, respectively and the pedigree was constructed with 7 generations.
For carcass and meat quality traits, data were collected from 1,953 pigs which were butchered at an abattoir in Namwon, but 11 incomplete records were deleted and only records from 1,942 pigs were used for the analysis and a total of 12 traits including (1) CW, (2) backfat thickness (BF), (3) pH value after 45 minutes (pH45m), (4) pH value after 24 hours (pH24h), (5) lightness in meat color (LMC), (6) redness in meat color (RMC), (7) yellowness in meat color (YMC), (8) moisture holding capacity (MHC), (9) drip loss (DL), (10) cooking loss (CL), (11) fat content (FC), and (12) shear force value (SH) were measured.
The CW (kg) and BF (mm) were measured at the parts of loins (Longissimus dorsi, LD) on the left side of the cold carcasses and used to determine meat quality parameters. The values for pH45m and pH24h were measured from the time of slaughter after inserting a pH meter (Orion 2 Star, Thermo Scientific, Waltham, MA, USA) directly into the muscle. The meat color values were measured using a colorimeter (CR-410, Minolta Co., Osaka, Japan) which was calibrated against a white reference tile plate (L* = 89.2, a* = 0.921, b* = 0.783). The color values for L* (lightness, LMC), a* (redness, RMC), and b* (yellowness, YMC) were obtained from the inside cuts of loins after 30 minutes blooming at room temperature. Centrifugation was used to measure MHC. Minced meat samples (5 g) were placed into a centrifuge tube with a filter paper (No. 4, Whatman International Ltd., Maidston, UK), and centrifuged at 3,000├Śg for 10 minutes. The MHC was calculated as the remaining moisture in the meat sample on the basis of the moisture content of the original meat sample. DL was measured as the percentage weight loss of a standardized (3├Ś3├Ś3 cm) meat sample placed in a sealed petri-dish at 4┬░C after 48 hours of storage. For the measurement of CL, the percentage weight loss of a standardized (3├Ś3├Ś3 cm) meat sample was measured after cooking in an electric grill with double pans (Nova EMG-533, 1,400 W, Evergreen Enterprise, Yongin, Korea) for 90 seconds, until the internal temperature of the meat sample reached 72┬░C. The parts of loins (3├Ś3├Ś2 cm) were cut and after installing a texture analyzer (TA-XT2, Stable Micro System Ltd., Godalming, UK) with a Warner-Bratzler blade, SH was measured with the blade cutting the muscle fibres at a right angle. Test and pre-test speeds were set at 2.0 mm/s and post-test speeds were set at 5.0 mm/s. The crude FCs were extracted according to the method described by Folch et al. (1957).
The analytical animal models (1 and 2) used for estimating the genetic parameters and expected breeding values (EBVs) were as follows:
Where, yijklm = reproductive traits, ╬╝ = overall mean, fi = fixed effect of the ith farrowed year-month-week (i = 1, 2, ŌĆ”, 276), lj = fixed effect of the jth parity (j = 1, 2, ŌĆ”, 8 more than), bk = random effect of the kth sire (k = 1, 2, ŌĆ”, 53), al = random additive genetic effect of the lth animal (l = 1, 2, ŌĆ”, 994), pl = permanent environmental effect of individual animals (l = 1, 2, ŌĆ”, 781) and eijklm = residual random effect.
Where, yijkl = carcass and meat quality traits, ╬╝ = overall mean, si = fixed effect of the ith sex (i = 1, 2), mj = fixed effect of the jth year-month of birth (j = 1, 2, ŌĆ”, 37), ak = random additive genetic effect of the kth animal (k = 1, 2, ŌĆ”, 2,487) and eijkl = residual random effect. Var(a) = A ŌŖŚ Žā2a, Var(pe) = I ŌŖŚ Žā2pe, and Var(e) = I ŌŖŚ Žā2e, where A = numerator relationship matrix and I = identity matrix. Basic statistical analysis was performed using SAS ver. 9.2 (SAS Institute Inc., Cary, NC, USA) and genetic parameters and EBVs for traits were estimated using VCE ver. 6.0 (Institute of Farm Animal Genetics, Friedrich Loeffler Institute, Neustadt, Germany) and PEST program (Institute of Animal Husbandry and Animal Behaviour, Federal Agricultural Research Centre, Neustadt, Germany). The genetic correlations between reproductive and meat quality traits were estimated using the EBVs of sows with progeny records of carcass and meat quality.
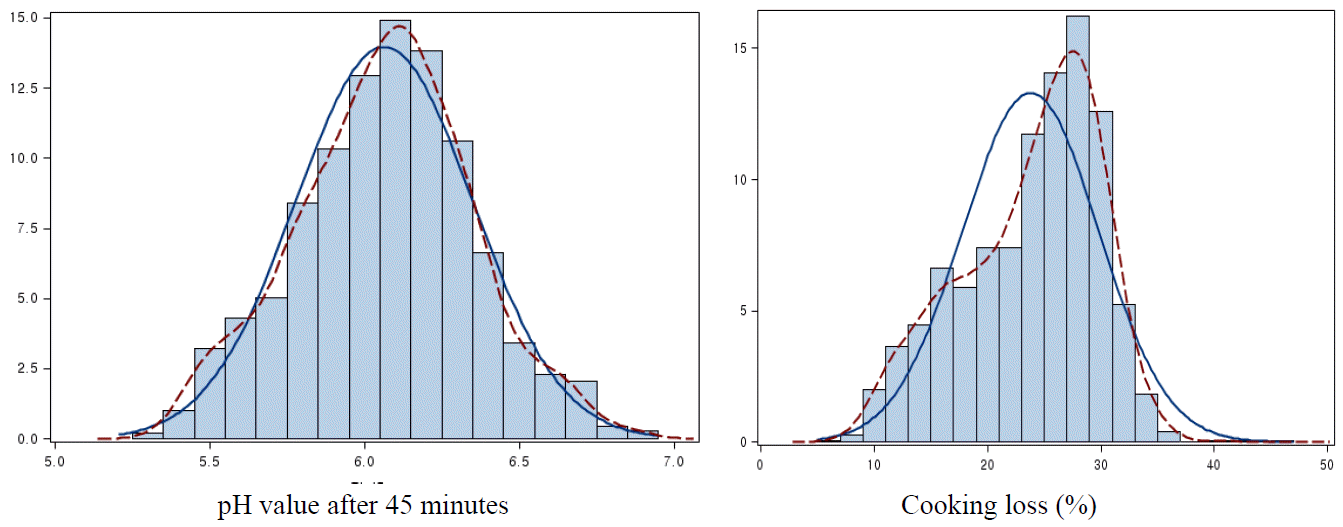
Basic data for the major economic traits of Berkshire pigs including reproductive, carcass and meat quality traits are shown in Table 1. The means for TNB and NBA were 8.65┬▒2.95 and 7.65┬▒2.58, respectively. The means for CW and BF were 86.62┬▒5.67 kg and 25.11┬▒5.23 mm, respectively and means for pH45m and pH24h, which are criteria for cadaveric metabolic rate, were 6.06┬▒0.29 and 5.78┬▒0.22, respectively. The means for LMC, RMC, and YMC, which are the evaluation indicators for abnormal meat, were 49.05┬▒3.18, 8.47┬▒4.34, and 3.59┬▒1.48, respectively. The means for MHC, DL, CL, and FC were 57.90┬▒11.10%, 4.14┬▒2.07%, 23.80┬▒6.00%, and 2.64┬▒1.06%, respectively and SH which is the indicator of tenderness was 2.82┬▒0.76 kg. Distributions of pH45m and CL which were considered as the most important traits for the genetic improvement in Dasan Pig Breeding Farm are shown in Figure 1. The pH24h showed a normal distribution, however, the CL was not normally distributed (p<0.01) by Shapiro Wilk test (SAS).
Heritability estimates for TNB and NBA in Berkshire were 0.07 and 0.06, respectively (Table 2), which were similar to those of Su et al. (2007) who reported the heritability of TNB in Landrace (0.08) and Yorkshire (0.05), and those of Chen et al. (2003a) in Landrace (0.08), Yorkshire (0.10), and Duroc (0.09). Generally, heritability for litter size seems to be low although it depends on the genetic makeup in a population (Arango et al., 2005). Repeatability estimates for TNB and NBA were 0.16 and 0.18, respectively and the sire effect was estimated as 1% of total variation, which was lower than that (5%) of Woodward et al. (1993), while genetic correlation between TNB and NBA was very high (0.988).
Heritabilities for carcass and meat quality traits are shown in Table 3. Heritability estimates for CW and BF were 0.37 and 0.57, respectively, which were similar to results by Tomiyama et al. (2011) who reported 0.54 for BF at finish and 0.32 for CW in a Japanese Berkshire population (n = 4,773). For other breeds, it was reported that the heritability for BF ranged from 0.47 to 0.56 (Bryner et al., 1992; Li and Kennedy, 1994; te Pas and Soumillion, 1999; Chen et al. 2002; Johnson and Nugent, 2003), which was similar to our results.
Heritability estimates for pH45m and pH24h, which are criteria for cadaveric metabolic rate, were 0.48 and 0.15, respectively. The heritability decreased sharply by the measurement time probably due to environmental factors such as storage temperature and management. The heritability estimates for pH24h by Sellier (1998), Hermesch et al. (2000), Suzuki et al. (2005), and Gaya et al. (2006) were 0.21, 0.07, 0.14, and 0.37, respectively. Since the pH45m and pH24h are the standards for muscle quality evaluation, they will be very important traits for meat quality improvement if we can obtain precise measurements. The heritability estimates for LMC was 0.19, which was lower than that (0.28) of Sellier (1998) and that (0.29) of Gaya et al. (2006). The heritability estimates for RMC and YMC were 0.36 and 0.28, respectively, which were higher than those (0.25 and 0.16) of Gaya et al. (2006). The heritability estimates for MHC, DL, and CL were 0.21, 0.33, and 0.45, respectively. It was reported that the heritability estimate for DL was 0.23 (Hermesch et al., 2000) or 0.25 (Gaya et al., 2006) and the heritability estimates for MHC and CL were in the range of 0 to 0.63 (Hovenier et al., 1993) and 0 to 0.51 (Sellier, 1998). The heritability estimate for FC was 0.43, which was similar to estimates obtained by Knapp et al. (1997; 0.42), Larzul et al. (1997; 0.44), and Hermesch et al. (2000; 0.35). The heritability estimates for SH was 0.39, which was lower than that by Suzuki et al. (2005; 0.45), but higher than that by Gaya et al. (2006; 0.22). These differences of estimated genetic parameters by other researches can be cause by the model, measurement and size of data used in the analysis.
The estimate for genetic correlation coefficient between CW and BF was 0.27, which was lower than that (0.42) obtained by Tomiyama et al. (2011) and the estimates for genetic correlation coefficients between CW and meat quality traits were in the range of ŌłÆ0.36 to 0.29 (Table 4). The genetic correlation coefficient between BF and FC was positive (0.27), while the genetic correlation coefficients between BF and CL and between BF and SH were negative (ŌłÆ0.28 and ŌłÆ0.30). The genetic correlation coefficients between pH24h and meat color traits were in the rage of ŌłÆ0.51 to ŌłÆ0.33, which were similar to those of Suzuki et al. (2005). The genetic correlation coefficients of pH24h for DL, CL, and SH were ŌłÆ0.41, 0.05, and ŌłÆ0.32, respectively.
The genetic correlation coefficients between DL and meat color traits were in the range of 0.34 to 0.53, and those between CL and meat color traits were in the range of 0.11 to 0.68, showing positive relationship.
Genetic correlation coefficient between DL and CL was 0.20. A higher value for genetic correlation (0.66) was reported by Sellier (1998), but Suzuki et al. (2005) reported a low genetic correlation coefficient (0.04) between DL and CL when measured by the same method used in this experiment. Genetic correlation coefficients between SH and FC was ŌłÆ0.17, which was similar to that (ŌłÆ0.20) by Suzuki (2005). Genetic correlation coefficients between SH and DL and between SH and CL were 0.05 and 0.44, respectively, and positively correlated. The estimates for genetic correlation coefficients between reproductive and meat quality traits were very low or zero (Table 5), which were similar to previous reports (Ducos and Bidanel, 1996; Clutter and Brascamp, 1998; Chen et al., 2003b; Holm et al., 2004). However, the estimates for genetic correlation coefficients between reproductive traits and DL and CL were in the range of 0.12 to 0.17, and ŌłÆ0.14 to ŌłÆ0.12, respectively, which were quite different from those of other meat quality traits, suggesting that DL and CL may have different water retention mechanisms.
Since carcass and meat quality traits have medium to high heritability, these valuable traits will respond to selection more quickly than reproductive traits since the heritability estimates for these traits are below 10%. Furthermore, the evaluation of meat quality, is very difficult to apply on the spot since various physical and chemical factors should be evaluated. Therefore, it seems to be practical to put emphasis on measuring pH value and backfat thickness, which are measurable on the spot, and then run parallel with selection for litter size. To overcome weaknesses of this study, the use of genomic selection would be desirable after establishing reference population for meat quality traits.
ACKNOWLEDGMENTS
This work was supported by a grant from the Next Generation BioGreen 21 Program (PJ01111502), Rural Development Administration, Republic of Korea.
Table┬Ā1
Means, standard deviations (SD) and skewness for reproductive (n = 2,511), carcass and meat quality traits (n = 1,942)
Table┬Ā2
Additive genetic (Žā2a), permanent environmental (Žā2pe), boar (Žā2b) and error variance (Žā2e) components, heritabilities (h2) and standard errors (SE), repeatabilities and phenotypic and genetic correlations for reproductive traits
| Traits | Žā2a | Žā2pe | Žā2b | Žā2e | h2┬▒SE | r | (1) TNB | (2) NBA |
|---|---|---|---|---|---|---|---|---|
| (1) TNB | 0.61 | 0.75 | 0.05 | 7.11 | 0.07┬▒0.03 | 0.16 | 0.916a | |
| (2) NBA | 0.44 | 0.70 | 0.04 | 5.31 | 0.06┬▒0.03 | 0.18 | 0.988b |
Table┬Ā3
Additive genetic (Žā2a) and error variance (Žā2e) components, and heritabilities (h2) and standard errors (SE) for carcass and meat quality traits
Table┬Ā4
Genetic and phenotypic correlations among carcass and meat quality traits
CW, arcass weight; BF, Back fat thickness; pH45m, pH value after 45 minutes; pH24h, pH value after 24 hours; LMC, lightness in meat color; RMC, redness in meat color; YMC, yellowness in meat color; MHC, moisture holding ability; DL, drip loss; CL, cooking loss; FC, fat content; SH, shear force value.
Table┬Ā5
Genetic correlations between carcass and meat quality traits and reproductive traits (pairs = 308)
| Traits | TNB | NBA |
|---|---|---|
| Carcass weight | ŌłÆ0.09NS | ŌłÆ0.05NS |
| Back fat thickness | ŌłÆ0.11NS | ŌłÆ0.12* |
| pH value after 45 minutes | ŌłÆ0.04NS | ŌłÆ0.03NS |
| pH value after 24 hours | 0.12* | 0.07NS |
| Lightness in meat color | ŌłÆ0.06NS | ŌłÆ0.03NS |
| Redness in meat color | ŌłÆ0.01NS | 0.01NS |
| Yellowness in meat color | ŌłÆ0.14* | ŌłÆ0.10NS |
| Moisture holding ability | 0.00NS | 0.03NS |
| Drip loss | 0.12* | 0.17** |
| Cooking loss | ŌłÆ0.12* | ŌłÆ0.14* |
| Shear force value | ŌłÆ0.02NS | ŌłÆ0.04NS |
| Fat | ŌłÆ0.07NS | ŌłÆ0.07NS |
REFERENCES
Arango J, Misztal I, Tsuruta S, Culbertson M, Herring W. 2005. Threshold-linear estimation of genetic parameters for farrowing mortality, litter size, and test performance of Large White sows. J Anim Sci 83:499ŌĆō506.


Bonneau M, Lebret B. 2010. Production systems and influence on eating quality of pork. Meat Sci 84:293ŌĆō300.


Bryner SM, Mabry JW, Bertrand JK, Benyshek LL, Kriese LA. 1992. Estimation of direct and maternal heritability and genetic correlation for backfat and growth rate in swine using data from centrally tested Yorkshire boars. J Anim Sci 70:1755ŌĆō1759.


Chen P, Baas TJ, Mabry JW, Dekkers JC, Koehler KJ. 2002. Genetic parameters and trends for lean growth rate and its comvonents in U.S. Yorkshire, Duroc, Hampshire, and Landrace pigs. J Anim Sci 80:2062ŌĆō2070.


Chen P, Baas TJ, Mabry JW, Koehler KJ, Dekkers JC. 2003a. Genetic parameters and trends for litter traits in U.S. Yorkshire, Duroc, Hampshire, and Landrace pigs. J Anim Sci 81:46ŌĆō53.


Chen P, Baas TJ, Mabry JW, Koehler KJ. 2003b. Genetic correlations between lean growth and litter traits in U.S. Yorkshire, Duroc, Hampshire, and Landrace pigs. J Anim Sci 81:1700ŌĆō1705.


Clutter AC, Brascamp EW. 1998. Genetics of performance traits. The Genetics of the Pig. Rothschild MF, Rubinsky A, editorsCAB International; New York, USA: p. 472ŌĆō462.

Ducos A, Bidanel JP. 1996. Genetic correlations between production and reproductive traits measured on the farm, in the Large White and French Landrace pig breeds. J Anim Breed Genet 113:493ŌĆō504.

Folch J, Lees M, Sloane Stanley GH. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 226:497ŌĆō509.


Gaya LG, Ferraz JBS, Balieiro JCC, Mattos EC, Costa AMMA, Michelan Filho T, Fel├Łcio AM, Rosa AF, Mour├Żo GB, Eler JP, Silva MEB, Queir├│z L, Af├Īz ALM, Longo NM, Garavazo BR, Nakashima SH. 2006. Heritability estimates for meat quality traits in a male Broiler line. In : Paper presented at the 8th World Congress on Genetics Applied to Livestock Production; Belo Horizonte, Brasil.
Hermesch S, Luxford BG, Graser HU. 2000. Genetic parameters for lean meat yield, meat quality, reproduction and feed efficiency traits for Australian pigs: 1. Description of traits and heritability estimates. Livest Prod Sci 65:239ŌĆō248.

Holm B, Bakken M, Klemetsdal G, Vangen O. 2004. Genetic correlations between reproduction and production traits in swine. J Anim Sci 82:3458ŌĆō3464.


Hovenier R, Kanis E, van Asseldonk Th, Westerink NG. 1993. Breeding for pig meat quality in halothane negative populations: A review. Pig News Inf 14:17NŌĆō25N.
Johnson ZB, Nugent RA. 2003. Heritability of body length and measures of body density and their relationship to backfat thickness and loin muscle area in swine. J Anim Sci 81:1943ŌĆō1949.


Knapp P, Willam A, S├Člkner J. 1997. Genetic parameters for lean meat content and meat quality traits in different pig breeds. Livest Prob Sci 52:69ŌĆō73.

Larzul C, Lefaucheur L, Ecolan P, Gogu├® J, Talmant A, Sellier P, Le Roy P, Monin G. 1997. Phenotypic and genetic parameters for longissimus muscle fiber characteristics in relation to growth, carcass, and meat quality traits in large white pigs. J Anim Sci 75:3126ŌĆō3137.


Li X, Kennedy BW. 1994. Genetic parameters for growth rate and backfat in Canadian Yorkshire, Landrace, Duroc, and Hampshire pigs. J Anim Sci 72:1450ŌĆō1454.


Luo W, Cheng D, Chen S, Wang L, Li Y, Ma X, Song X, Liu X, Li W, Liang J, Yan H, Zhao K, Wang C, Wang L, Zhang L. 2012. Genome-wide association analysis of meat quality traits in a porcine Large White├ŚMinzhu intercross population. Int J Biol Sci 8:580ŌĆō595.



Newcom DW, Baas TJ, Mabry JW, Goodwin RN. 2002. Genetic parameters for pork carcass components. J Anim Sci 80:3099ŌĆō3106.


SAS Institute Inc. 2008. SAS/STAT 92 UserŌĆÖs Guide. SAS Institute Inc; Cary, NC, USA:
Sellier P. 1998. Genetic of meat and carcass traits. The Genetics of the Pigs. Rothschild MF, Rubinsky A, editorsCAB International; New York, USA: p. 465ŌĆō510.
Su G, Lund MS, Sorensen D. 2007. Selection for litter size at day five to improve litter size at weaning and piglet survival rate. J Anim Sci 85:1385ŌĆō1392.


Suzuki K, Irie M, Kadowaki H, Shibata T, Kumagai M, Nishida A. 2005. Genetic parameter estimates of meat quality traits in Duroc pigs selected for average daily gain, longissimus muscle area, backfat thickness, and intramuscular fat content. J Anim Sci 83:2058ŌĆō2065.


Pas MF, Soumillion A, Harders FL, Verburg FJ, van den Bosch TJ, Galesloot P, Meuwissen TH. 1999. Influences of myogenin genotypes on birth weight, growth rate, carcass weight, backfat thickness, and lean weight of pigs. J Anim Sci 77:2352ŌĆō2356.


Tomiyama M, Kanetani T, Tatsukawa Y, Mori H, Oikawa T. 2011. Genetic relationships and expected responses for genetic improvement of carcass traits of Berkshire pigs. Sci Agric (Piracicaba, Braz) 68:594ŌĆō597.






 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Full text via PMC
Full text via PMC Download Citation
Download Citation Print
Print





