 |
 |
| Anim Biosci > Volume 35(1); 2022 > Article |
|
Abstract
Objective
FKBP prolyl isomerase 5 (FKBP5) has been shown to play an important role in metabolically active tissues such as skeletal muscle. However, the expression of FKBP5 in Muscovy duck tissues and its association with body weight are still unclear.
Methods
In this study, real-time quantitative polymerase chain reaction was used to detect the expression of FKBP5 in different tissues of Muscovy duck at different growth stages. Further, single nucleotide polymorphisms (SNPs) were detected in the exon region of FKBP5 and were combined analyzed with the body weight of 334 Muscovy ducks.
Results
FKBP5 was highly expressed in various tissues of Muscovy duck at days 17, 19, 21, 24, and 27 of embryonic development. In addition, the expression of FKBP5 in the tissues of female adult Muscovy ducks was higher than that of male Muscovy ducks. Besides, an association analysis indicated that 3 SNPs were related to body weight trait. At the g.4819252 A>G, the body weight of AG genotype was significantly higher than that of the AA and the GG genotype. At the g.4821390 G>A, the genotype GA was extremely significantly related to body weight. At the g.4830622 T>G, the body weight of TT was significantly higher than GG and TG.
Skeletal muscle, which accounts for 50% to 60% of the total weight, is one of the major tissues involved in regulating metabolism, movement, and strength [1]. In addition, skeletal muscle forms a network that regulates the function of distal organs and skeletal muscle itself by releasing various types of actin [2]. In livestock production, muscle yield is one of the most important factors in determining the economic value [3]. Skeletal muscle mass and fiber size are regulated in response to changes in animal activity, disease and aging through many genes, transcription factors or non-coding RNAs (lncRNA or circRNA) [4]. Skeletal muscle development is a rigorous procedural process that goes through several periods, i) Differentiation of muscle precursor cells from the somite; ii) The muscle precursor cells proliferate and differentiate into myoblasts; iii) Myoblasts fusion into myotubes; iv) Myotubes form muscle fibers [5,6]. Besides, at the late embryonic stage, myoblasts stop the cell cycle and fuse to form myotubes.
FK506 binding proteins (FKBPs) are multi-functional proteins that are highly conserved between species. They are abundantly expressed in cells and belong to members of the immunoaffinity protein family [7]. The molecular weight of FKBPs range from 12 kDa to 135 kDa. FKBP51, also known as FKBP5, has a molecular weight of 51 kDa, is involved in regulation of immune and basic protein folding and transport, and it is a member of the immunoaffinity protein family. The main research areas of FKBP51 include mental illness, anxiety, and depression [8,9], which may be due to the pivotal role of FKBP51 in the hypothalamic-pituitary-adrenal axis (HPA-axis) [10]. The HPA axis has a feedback control system that regulates the release of glucocorticoids [11], and FKBP51 has its ability to modulate glucocorticoid receptor (GR) sensitivity and HPA axis function [12,13]. FKBP51, as part of the heat shock protein 90 (Hsp90) steroid receptor complex, plays an important role in regulating the steroid hormone receptor (GR). Glucocorticoids induce expression of FKBP5 at mRNA and protein levels as part of an intracellular negative feedback loop that regulates GR activity [14]. Moreover, FKBP5 also plays an important role in metabolic regulation, such as inhibiting AKT [15] and mammalian target of rapamycin signal regulated kinases, and mediating autophagy [16], which is a catabolic process that maintains homeostasis, and is associated with a metabolic phenotype. FKBP5 is confirmed to be most expressed in human skeletal muscle and adipose tissue in all study tissues [17], and increased FKBP5 expression in omental adipose tissue is associated with insulin resistance [18]. In mice, the expression of FKBP5 in medial hypothalamus (the brain area that regulating satiety and body weight) is induced by a high-fat diet and is positively correlated with stress-induced weight gain [19]. Besides, compared to wild-type mice, mice lacking FKBP5 gene lose weight and are resistant to diet-induced obesity [20]. Some studies have believed that FKBP5 may affect body weight and metabolism through multiple pathways and actions [21]. Although the mechanisms of these effects remain unclear, studies to date have agreed that up-regulation of FKBP5 is associated with weight gain and negative metabolism.
FKBP5 is also involved in NF-κB and Akt signaling pathways and it regulates the development of osteoclasts, survival, and activation of bone resorption. Lu et al [22] found that the novel mutation (c.163G>C) in FKBP5 gene was associated with Paget’s disease of bone (PDB), and the mutation promoted osteoclast differentiation and bone resorption activity, both of which were major defect in PDB development. Hartmann et al [23] found that the excess body weight of carriers of the FKBP5 rs1360780 polymorphism T allele was nearly 20% less than that of homozygous carriers of the C allele, and overall weight reduced 10% at 26 weeks of follow-up. Yeo et al [24] showed that changes in the rs1360780 locus could influence the expression of FKBP5, which affected the expression of other glucocorticoid-regulated genes. Therefore, FKBP5 is speculated to be a potential gene that regulates muscle development.
Muscovy duck, also known as red-billed goose or musk duck, is native to the tropical regions of Central and South America. It was introduced to China more than 250 years ago. Muscovy duck has the advantages of large body size, rapid growth, resistance to rough feeding, easy fattening, delicious meat, and good liver performance. Currently, there are few reports on FKBP5 gene in Muscovy ducks. Therefore, the expression patterns of FKBP5 in Muscovy duck tissues at different growth stages were first studied, and then the association between Muscovy duck FKBP5 single nucleotide polymorphisms (SNPs) and body weight was used to determine whether FKBP5 could be used as a candidate gene for Muscovy duck growth.
All animal procedures were performed according to guidelines provided by the China Council on Animal Care, and the protocols were approved by the Experimental Animal Management Committee (EAMC) of Northwest A&F University.
The eggs of Muscovy duck with similar size and weight were incubated in accordance with the incubation procedure after disinfection. Eight embryo eggs were randomly selected at the 17 d (E17d), 19 d (E19d), 21 d (E21d), 24 d (E24d), 27 d (E27d), 31 d (E31d), and 34 d (E34d) during the incubation period, respectively. Heart, liver, lung, kidney, breast muscle, and leg muscle were separated for RNA extraction. Similarly, 6-month-old Muscovy ducks were randomly selected (4 males and 4 females), and the tissues containing heart, liver, spleen, lung, kidney, breast muscle, leg muscle and testis/ovary were quickly isolated for RNA extraction after slaughter. Besides, a total of 344 Muscovy ducks (280 days old and unrelated for at least three generations) were randomly selected for blood sampling. Body weight data of Muscovy duck was recorded for association studies. Adult Muscovy ducks were raised under the same environmental conditions and had free access to feed and water (Supplementary Table S1). All Muscovy ducks and eggs were purchased from Anda Farm in Tongguan County, Shaanxi Province, China.
RNA was isolated from different tissues of embryonic and 6-month-old Muscovy ducks using TRIzol Reagent (CWBIO, Taizhou, Jiangsu, China) according to the manufacturer’s instructions. The extracted RNA was detected by 1% agarose gel electrophoresis and NanoDrop 2000 (Thermo, Waltham, MA, USA) for quality and concentration, respectively. The first strand cDNA was synthesized from purified RNA by reverse transcription kit (abm, Richmond, BC, Canada) according to the manufacturer’s protocol.
Spanned exon primers were designed by primer premier 5.0 based on the mRNA sequence of duck FKBP5 gene (GenBank accession number: NC_040072.1) and β-actin gene (GenBank accession number: NC_040060.1) from GenBank (www.ncbi.nlm.nih.gov/) (Table 1). The relative expression of target gene was normalized against the internal control gene, β-actin. To determine the real-time quantitative polymerase chain reaction (qRT-PCR) efficiency of target and internal control genes, tenfold serial dilutions (10−1 to 10−5) of cDNA were produced and assayed in triplicate to yield standard curves. The identity of the amplified products was also confirmed by sequencing. The qRT-PCR reactions were performed in a total volume of 10 μL including 5 μL 2×TransStart Tip Green qPCR SuperMix (Transgen, Beijing, China), 0.2 μL of each primer (10 μM), 0.8 μL cDNA (400 ng/μL) and 3.8 μL ddH2O. Each cDNA sample was then analyzed in triplicate using EcoRT48 system (OSA, London, UK). The optimum thermal cycling conditions consisted of 95°C for 30 s, followed by 40 cycles of 95°C for 5 s, 60°C for 30 s, then 95°C for 15 s, 55°C for 15 s, 95°C for 15s. The relative expression level of FKBP5 was calculated by 2−ΔΔCt method. All data were subjected to one-way analysis of variance among different tissues.
Genomic DNA from Muscovy duck blood was extracted according to the phenol chloroform protocol, then DNA concentration and quality were measured by NanoDrop 2000 and 1% agarose gel electrophoresis, respectively. Finally, the genomic DNA was diluted to 80 ng/μL for storeage at −20°C. According to the nucleotide sequence of duck FKBP5 gene (GenBank accession number NC_040072.1), 9 pairs of PCR primers (Table 1) were designed using primer premier 5.0 software. Each amplification reaction was carried out in a final volume of 15 μL reaction mixture containing 7.5 μL 2×Taq PCR Mix (CWBIO, China), 0.8 μL genomic DNA, 0.4 μL of each primer (10 μM), 5.9 μL ddH2O. The reactions were performed with the protocol consisting of initial denaturation at 94°C for 5 min, followed by 34 cycles of 94°C for 30 s, 30 s at the annealing temperature, and 60 s at 72°C, ending with a final extension at 72°C for 10 min.
To determine the mutation sites in the Muscovy duck FKBP5 gene, 30 individual DNA samples were randomly selected to mix for a DNA pool. Subsequently, PCR amplifications were performed on two DNA pools and three random DNA individuals according to the above reaction conditions and reaction procedures. The target fragments were sequenced after verification by 1.5% agarose gel electrophoresis. Finally, the sequences were imported into the Chromas 1.62 for SNPs detection.
According to the location of mutation site, the restriction enzymes were selected for genotyping (Table 1). The digested reaction was carried out in 10 μL reactions containing 6 μL PCR product, 0.2 μL restriction enzyme (Takara, Tokyo, Japan or Thermo Scientific, USA), 1 μL 10×Buffer, 2.8 μL ddH2O, then it was reacted for 3 h after well mixed. The digested fragments were detected by 3% agarose gel electrophoresis.
The allele frequency, genotype frequency, genotype frequency distribution, χ2 independence test, and genetic variation index, the gene heterozygosity (He), the effective allele numbers (Ne) and the relationship between the genotypes and the body weight of Muscovy duck were statistically analyzed. And polymorphism information content (PIC) was calculated by an online tool (www.msrcall.com). Association analysis of SNP and body weight was done using SPSS 20.0. The haplotype analysis was performed using Haploview (http://analysis.biox.cn/myAnalysis.php) [25].
The following general linear model was used for association test: Yijk = μ+Mi+Gj+Fx+ eijk, where Yijk, the observed value; μ, the overall mean of individual traits; Mi, the fixed effect of slaughtering age; Gj, the fixed effect of genotype; Fx, the fixed effect of sex; eijk, the random error.
The expression levels of FKBP5 gene in Muscovy ducks at different hatching stages were detected by qRT-PCR (Figure 1). In heart of Muscovy duck, the relative expression of FKBP5 gene in E17d, E19d, E21d, E24d, and E27d was significant or extremely significantly higher than that of E31d (p<0.01 or p<0.05, Figure 1A). The qRT-PCR results showed that the relative expression trends of FKBP5 in liver and lung were similar to that in heart, that is, the expression of FKBP5 in E17d, E19d, E21d, E24d, and E27d were significant or extremely significantly higher than that of E31d (p<0.01 or p<0.05, Figure 1B, 1C). Besides, the expression trends in kidney and breast muscle were similar to leg muscle, where the expressions in kidney at the day 17, 19, 24, 27 of embryonic were extremely significantly higher than that of E31d (p<0.01, Figure 1D), and compared with the expression in E31d, FKBP5 expression was significantly or extremely significantly higher in breast muscle in E17d, E19d, E21d, E24d, and E27d (p<0.01 or p<0.05, Figure 1E). FKBP5 expressions in leg muscle in E17d, E19d, E21d, E24d, and E27d were significantly higher than that of E31d (p<0.01, Figure 1F). In all tissues, the expression levels of FKBP5 in E34d were lower than that in E31d, but the differences were not significant (p>0.05).
To determine the expression levels of duck FKBP5 gene in different genders, 6-month-old male and female Muscovy ducks were selected as research objects. The expression of FKBP5 gene in the tissues of female ducks was higher than male ducks’ (except liver), and in leg muscle of female ducks, the expression of FKBP5 gene was significantly higher than that of the male Muscovy ducks (p<0.05). In addition, the expression of FKBP5 gene in ovary was significantly higher than that in testis (p<0.01, Figure 2).
To verify the relationship between the SNP locus of FKBP5 gene and duck body weight, blood samples were collected from 344 Muscovy ducks for DNA extraction. Then 9 pairs of primers were designed to amplify the duck FKBP5 gene fragments (Figure 3). A total of 8 SNPs were identified by pool DNA or individual DNA sequencing, including g.4819189 C>T, g.4819220 T>C, g.4819252 A>G, g.4819303 C>T, g.4821390 G>A, g.4825015 C>T, g.4830197 T>C, and g.4830622 G>T (Figure 4), and the corresponding restriction enzymes are shown in Table 1. Eight SNP loci were detected by PCR-restriction fragment length polymorphism, as shown in Figure 5. In the sequence of S1, 4 mutation sites were found, and these SNPs which g. 4819189 C>T, g. 4819220 T>C, g. 4819252 A>G, g. 4819303 C>T were genotyped by the restriction enzyme Hha I (resulted in three banding patterns: digested genotypes CC, CT, and TT), Sac II (digested genotypes TT, TC, and CC), Not I (digested genotypes AA, AG, and GG) and Nco I (digested genotypes CC, CT, and TT), respectively. The PCR fragment of S4 in FKBP5 had a restriction site, which g. 4821390 G>A was cut into GG, GA, and AA by Hha I. The sequence of S5 (g. 4825015 C>T) was cleaved into TT, TC, and CC by Sma I. Product of S8 (g. 4830197 T>C) was cut into CC, CT, and TT by Hha I. Besides, the mutation site of S9, which g. 4830622 C>T was digested into the genotype TT, CT, and CC by Ban I.
The SNP allele and genotype frequency of FKBP5 in Muscovy duck are shown in Table 2, the results showed a consensus that the C allele was dominant in 3 SNPs of g.4819189 C>T, g.4819303 C>T and g.4825015 C>T, which were 71.15%, 50.60%, and 56.85%, respectively. The A allele in g.4819252 A>G and g.4821390 G>A was dominant, accounting for 58.80% and 53.70%, respectively. Additionally, the frequencies of allele T were 81.80% (g.4819220 T > C), 78.20% (g.4830197 T>C) and 75.55% (g.4830622 G>T) in the other three SNP loci, respectively. The χ2 test results showed that all SNP sites were consistent with Hardy-Weinberg equilibrium. The diversity parameter He of Muscovy duck was between 0.2986 and 0.5891, and the Ne values were between 1.4256 and 1.9997. In addition, the values of PIC possessed intermediate polymorphism (0.25<PIC<0.5), which were between 0.2540 and 0.3750.
Relationships between the 8 SNPs of FKBP5 and body weight trait were examined in 334 Muscovy ducks (Table 3). At the g.4819252 A>G (located in intron region), the genotype-AG individuals had greater body weight than the AA and the GG individuals (p<0.05), and the weight of GG genotype of Muscovy duck was significantly higher than that of the AA (p<0.05). At the g.4821390 G>A (located in intron region), genotype GA was extremely significantly related to body weight (p<0.01). Besides, at the g.4830622 T>G (located in 3′ UTR region), the TT type had a body weight that was significantly higher than the GG and the TG individuals (p<0.05), indicating that genotype TT was related to the body weight of Muscovy duck. In addition, other SNP loci were not associated with Muscovy duck weight (p>0.05). Haplotypes from the 344 individuals were analyzed using Haploview, and 8 haplotypes were found in g.4819252 A>G, g.4821390 G>A and g.4830622 T>G. AAG was the most common haplotype, with a frequency of 22.9%; next came AGG, GAG, and GGG, occurring with frequencies of 21.7%, 20.1%, and 10.9%, respectively (Table 4).
Skeletal muscle plays a key role in the movement of the body to maintain vital activity. At the same time, skeletal muscle affects the energy metabolism of the whole body by regulating such as the fine energy production and consumption system. In animal husbandry, muscle production and muscle growth rate are factors that affect the important economic value of livestock and poultry. In addition to being closely related to breed and feeding condition [26], skeletal muscle development of animals is also associated with muscle-related regulatory genes. To date, there have been many studies on gene expression related to muscle development [27,28]. FKBP5 gene is involved in cellular actions including regulation of cell proliferation, autophagy, osteoclast formation and insulin resistance in adipose tissue. In mice, FKBP5 increased the mass of skeletal muscle, which may be through the enhancement of muscle protein synthesis and myotube differentiation, as well as inhibition of muscle protein degradation [29]. Therefore, we aimed to explore the expression of FKBP5 in Muscovy duck tissues, as well as the relationship between FKBP5 SNPs and body weight. In this study, there was a significant difference in FKBP5 gene expression between the embryonic and adult Muscovy duck tissues. In addition, a significant relationship between SNPs and body weight characteristic in Muscovy duck population was found.
In the middle and late stages of embryo development, the tissues of Muscovy ducks developed rapidly. In our study, from the 17 d prior to hatching, the tissues were separated every 2 to 4 days, and the expression level of FKBP5 was analyzed by qRT-PCR. Results of qRT-PCR indicated that FKBP5 mRNA was widely expressed in different tissues of Muscovy duck. In E17d, E19d, E21d, E24d and E27d, the expression levels of FKBP5 gene in Muscovy duck heart, liver, lung, breast muscle and leg muscle were significantly or extremely significantly higher than that of E31d (p<0.05 or p<0.01), while the expressions of FKBP5 gene in kidney in E17d, E19d, E24d, and E27d were extremely significantly higher than those of the 31 d (p<0.01). In all tissues, the expressions of FKBP5 gene in E34d were lower than that of E31d, but the differences were not statistically significant (p>0.05). It was indicated that the expression level of FKBP5 had been reduced to a low level at the end of the embryonic period. In particular, in breast muscle of Muscovy duck, FKBP5 was highly expressed in the middle stage of hatching, and it reached the highest point in E19d. It was speculated that the duck’s breast muscle was developing rapidly at this time. With the extension of incubation time, the expression of FKBP5 gene gradually decreased to the lowest level after E31d. It was suspected that the duck’s breast muscle had basically developed. Similarly, the expression of FKBP5 in leg muscle was highly expressed before E27d, and the expression was gradually reduced after E31d. The results showed that the Muscovy duck’s leg muscle developed rapidly before the 27 d of hatching, and the development of leg muscle was nearing completion after 31 d of the incubation. FKBP5 plays an important role in regulating the individual metabolism [15,30]. Compared with other tissues, the high expression level of FKBP5 in metabolically active tissues (such as skeletal muscle and adipose tissue) further supports the role of FKBP5 in systemic metabolism [17]. In the embryonic stage of Muscovy duck, the tissues such as heart, liver, lung, kidney, and skeletal muscle developed rapidly and metabolized vigorously, which may be one of the reasons for the high expression of FKBP5. Therefore, FKBP5 may promote the growth and development of Muscovy duck, especially skeletal muscle, at the middle and late stages of embryo development. Interestingly, in the tissues of adult female Muscovy ducks, the expressions of FKBP5 were always higher than that of male Muscovy ducks (except liver). More importantly, the expression of FKBP5 in leg muscle of female Muscovy ducks was significantly higher than male ducks. In addition, the gene expression in ovary was significantly higher than that of testis. It could be explained by the fact that FKBP51, as part of the heat shock protein 90 steroid receptor complex, plays an important role in regulating the steroid hormone receptor (GR), which involves a wide range of physiological metabolic processes including immune, cardiovascular, reproductive, neurological, and metabolic systems [31]. Due to egg production and other factors (such as follicle development), the physiology and metabolism of female ducks are higher than that of males [32]. Therefore, the expressions of FKBP5 in female Muscovy duck tissues were higher than that in male Muscovy duck.
So far, there were few studies about the association analysis between SNPs of FKBP5 and body weight. Therefore, we mainly focused on mutations of FKBP5 in Muscovy duck and studied whether mutations of FKBP5 affected the body weight. By sequencing the PCR products, 8 SNP sites were found. Then enzyme digestion on these SNP sites was performed, and statistical analysis was conducted. The χ2 test results showed that all SNP loci conformed to the Hardy-Weinberg equilibrium law. It meant that the frequency of each allele was stable. In other words, the genetic balance was maintained. The genetic parameters He and Ne of the SNP loci in Muscovy duck population ranged from 0.2986 to 0.5891 and 1.4256 to 1.9997, respectively. PIC value represented the level of genetic diversity (PIC value <0.25, low genetic diversity; 0.25<PIC value<0.50, intermediate genetic diversity; and PIC value >0.50, high genetic diversity), the values of PIC were in the range of 0.2540 to 0.3750, which were a moderate polymorphic range, indicating that each SNP locus was more polymorphic in Muscovy duck population.
Finally, 3 SNP loci (g.4819252 A>G, g.4821390 G>A, and g.4830622 T>G) were found in Muscovy duck FKBP5 gene related to body weight, and haplotype results showed that the frequencies of AAG, AGG, GAG, and GGG were higher. Because muscle accounts for 50% to 60% of body weight, we believe that these SNP sites may regulate muscle development and increase muscle mass. Specifically, genotype AG of g.4819252 A>G, genotype GA of g.4821390 G>A and genotype TT of g.4830622 T>G were highly correlated with Muscovy duck weight. Notably, g.4819252 A>G and g.4821390 G>A are in the intron of FKBP5 gene. The transcription efficiency of genome may be directly affected by the base mutation in intron region of gene. Six intron SNPs of ECHS1 were significantly associated with milk fatty acids in dairy cows [33]. Cao et al [34] found that SNP sites in introns may change the formation and occurrence of the spliceosome, and ultimately affect protein biosynthesis. In addition, g.4830622 T>G is in the 3′ UTR region of FKBP5, and the 3′UTR SNP can regulate the post-transcriptional gene expression by affecting gene transcription activity, mRNA splicing, stability of mRNA and the binding of miRNA to target mRNA [35]. Ju et al [36] suggested that the SNP g.18475 A>G in 3′UTR of neutrophil cytosolic factor 4 (NCF4) was associated with mastitis susceptibility in dairy cows. Therefore, our results suggested that FKBP5 could see potential use as a molecular marker in future breeding programs to increase growth rates of Muscovy ducks.
In this study, the expressions of FKBP5 gene were detected in embryonic and adult Muscovy duck tissues. The results showed that FKBP5 was significantly expressed in different tissues at the stages of embryonic development. In addition, the expressions of FKBP5 in adult female duck tissues were higher than that of male ducks. Subsequently, the correlation between Muscovy duck FKBP5 SNPs and body weight was studied. It was found that 3 SNPs (g.4819252 A>G, g.4821390 G>A, and g.4830622 T>G) were significantly associated with Muscovy duck weight. These findings indicated that FKBP5 could be used as a molecular marker for muscle development traits by early marker-assisted selection of Muscovy duck.
Notes
SUPPLEMENTARY MATERIAL
Supplementary file is available from: https://doi.org/10.5713/ab.20.0649Supplementary Table S1
The feed composition of adult Muscovy duck
Figure 1
Expression of FKBP5 gene at embryonic stages of Muscovy duck in various tissues. (A) Expression of FKBP5 gene in heart. (B) Expression of FKBP5 gene in liver. (C) Expression of FKBP5 gene in lung. (D) Expression of FKBP5 gene in kidney. (E) Expression of FKBP5 gene in breast muscle. (F) Expression of FKBP5 gene in leg muscle. The data were normalized to the expression of FKBP5 on day 31 of incubation and calculated by using 2−ΔΔCt. β-actin was measured as the reference gene. Columns are represented as mean±standard deviation for three independent experiments performed in triplicate. FKBP5, FKBP prolyl isomerase 5. * Indicates that the difference is significant (p<0.05), ** indicates that the difference is extremely significant (p<0.01).
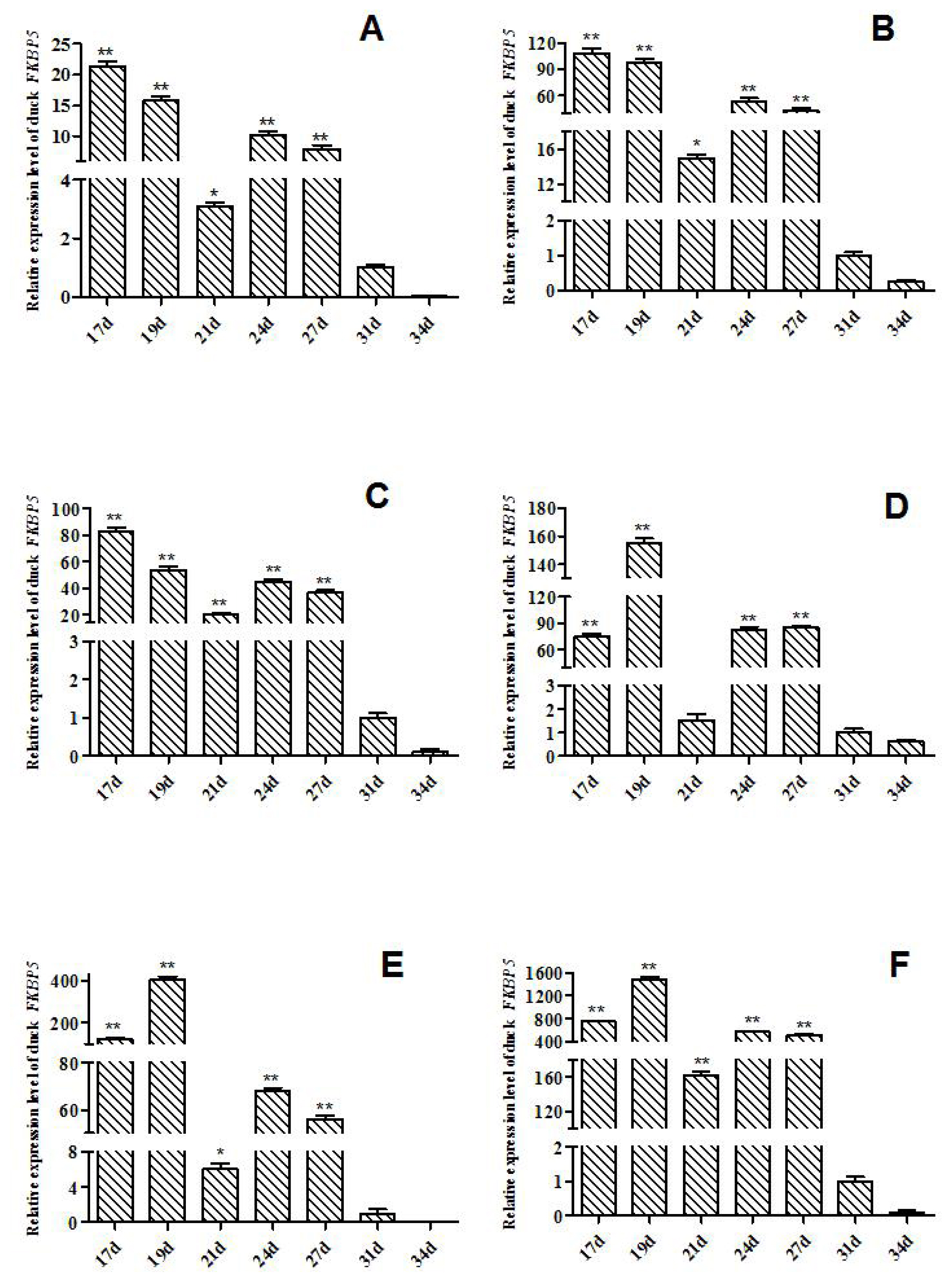
Figure 2
Expression of FKBP5 gene in various tissues of 6-month-old Muscovy duck. The data were normalized to the expression of FKBP5 in male Muscovy duck and calculated by using 2−ΔΔCt. β-actin was measured as the reference gene. Each Column represented the mean±standard deviation of three independent experiments which were performed in triplicate. FKBP5, FKBP prolyl isomerase 5. * Indicates that the difference is significant (p<0.05), ** indicates that the difference is extremely significant (p<0.01).

Figure 3
The amplification of FKBP5 gene. The fragments of S1–S9 were 634 bp, 609 bp, 777 bp, 541 bp, 717 bp, 655 bp, 387 bp, 803 bp, 648 bp, respectively. Lane M was DNA DM 2000 Marker. FKBP5, FKBP prolyl isomerase 5.
Figure 4
SNPs sequencing chromatogram of primer in FKBP5 gene. (A) SNP locu of g.4819189 C>T. (B) SNP locu of g.4819220 T>C. (C) SNP locu of g.4819252 A>G. (D) SNP locu of g.4819303 C>T. (E) SNP locus of g.4821390 G>A. (F) SNP locu of g.4825015 C>T. (G) SNP locu of g.4830197 T>C. (H) SNP locu of g.4830622 G>T. The Chromas 1.62 software was used to detect FKBP5 SNPs. SNPs, single nucleotide polymorphisms; FKBP5, FKBP prolyl isomerase 5.
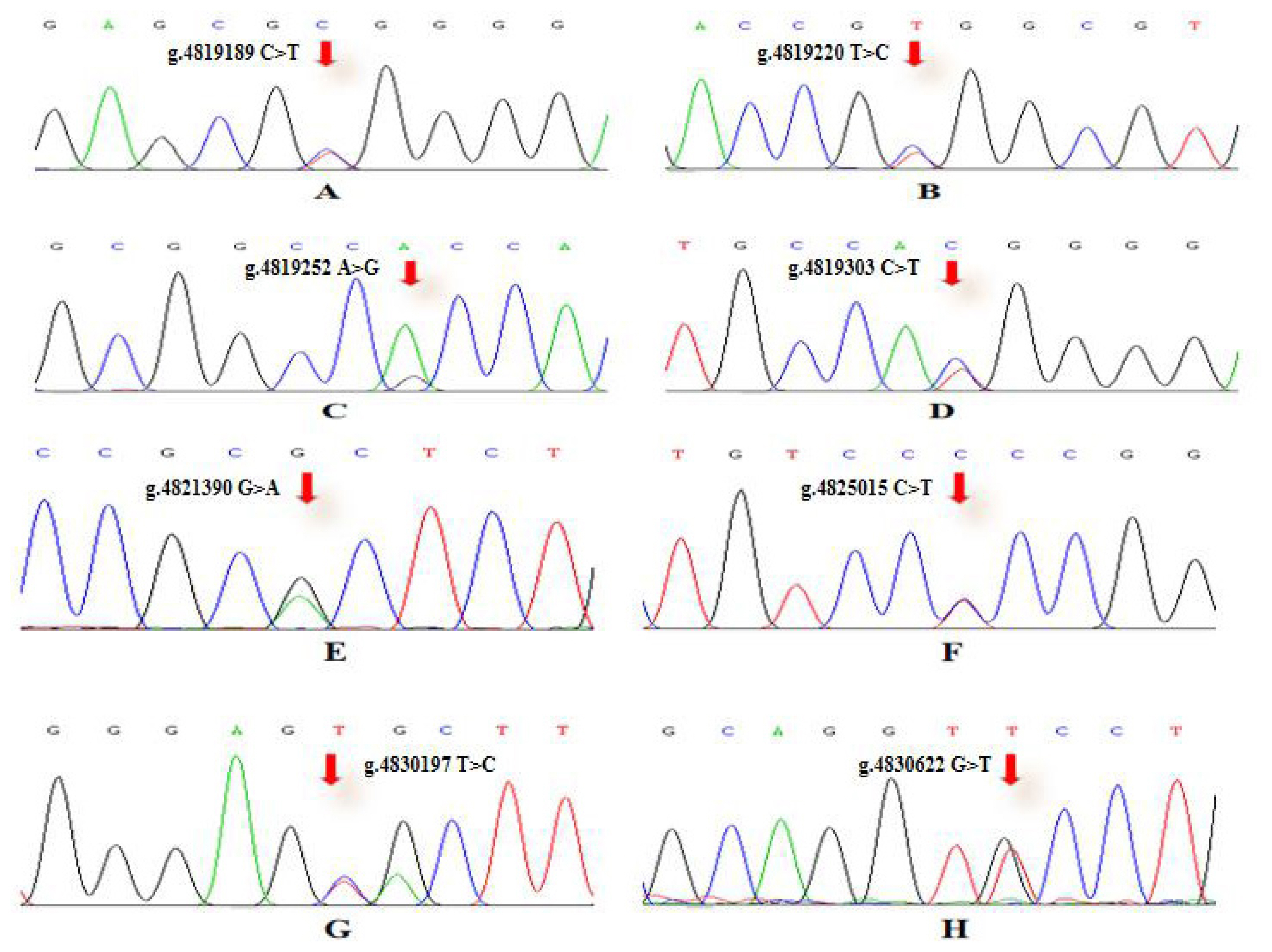
Figure 5
PCR-RFLP detection of the FKBP5 gene PCR products. (A) Digestion of g. 4819189 C> T site by Hha I. (B) Digestion of g.4819220 T>C site by Sac II. (C) Digestion of g.4819252 A>G site by Not I. (D) Digestion of g.4819303 C>T site by Nco I. (E) Digestion of g.4821390 G>A site by Hha I. (F) Digestion of g.4825015 C>T site by Sma I. (G) Digestion of g.4830197 T>C site by Hha I. (H) Digestion of g.4830622 G>T site by Ban I. Lane M was DNA marker I. PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism; FKBP5, FKBP prolyl isomerase 5.
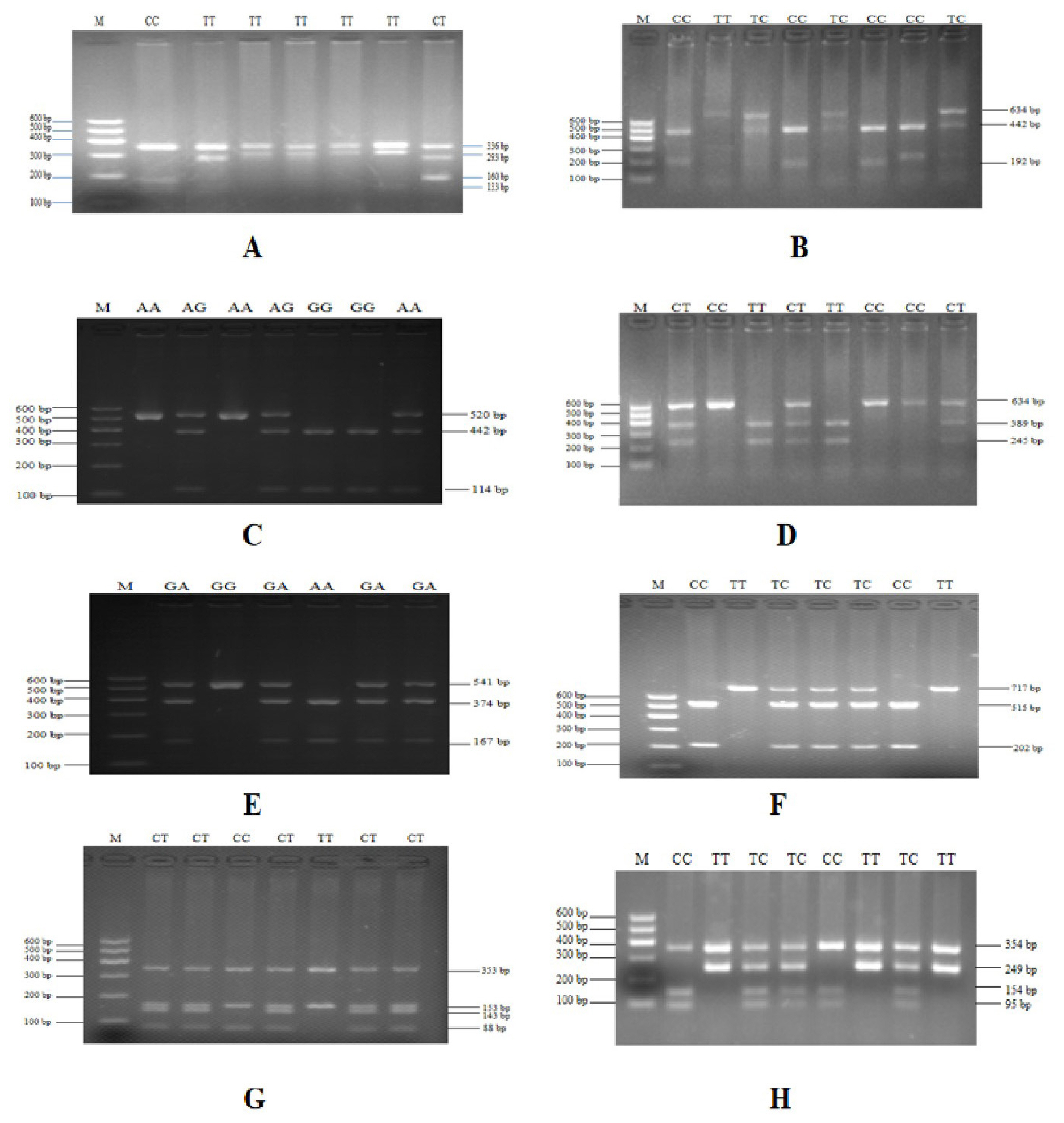
Table 1
qRT-PCR and PCR Primers for duck FKBP5 gene
| Name | Primers (5′→3′) | Fragment size (bp) | Restriction enzyme1),2) |
|---|---|---|---|
| RT-FKBP5 | Forward primer TCCAACGCCACCCTCTTC | 193 | |
| Reverse primer ACTTCACGTCCTTGCAGTCG | |||
| RT-β-actin | Forward primer CCCTGTATGCCTCTGGTCG | 194 | |
| Reverse primer CTCGGCTGTGGTGGTGAAG | |||
| S1 | Forward primer TCCAGCTTTTAACCCTTTCC | 634 | Hha I (336, 293, 160, 133, 5), Sac II (634, 442, 192) |
| Reverse primer CGTTTCACTACTCGCTTCTTGT | Not I (520, 442, 114), Nco I (634, 389, 245) | ||
| S2 | Forward primer CCTGCGAGTGAAATCTGC | 609 | None |
| Reverse primer GATGTCCCACGCCTTGAT | |||
| S3 | Forward primer GCACTACAAAGGCAAACTG | 777 | None |
| Reverse primer AGCACCTGAAGCCCAACA | |||
| S4 | Forward primer GTGGCTTCCCCGTTGTTT | 541 | Hha I (541, 374, 167) |
| Reverse primer GACGCAGGAGGGGTAAAT | |||
| S5 | Forward primer TAATCGCCGTGATAAACTCC | 717 | Sma I (717, 515, 202) |
| Reverse primer CCTTCCACCGAGGCTAAA | |||
| S6 | Forward primer AGCCCCTCAAACTCAACG | 655 | None |
| Reverse primer TTTCCAGCCAGGACACGA | |||
| S7 | Forward primer CTGGCGAACTCTGTCCTT | 387 | None |
| Reverse primer ACCTCAGCATCCTCAACCC | |||
| S8 | Forward primer TTGCCAGGACTCAAGGAC | 803 | Hha I (353, 153, 143, 88, 66, 55) |
| Reverse primer CCCAGCTAAGCGATAAAAC | |||
| S9 | Forward primer AGGCGTTTCTCATCCGTG | 648 | Ban I (354, 249, 154, 95, 45) |
| Reverse primer GCTGAGATCCCAAACAAC |
Table 2
Allele and genotype frequencies of FKBP5 SNP locus of Muscovy duck
Table 3
Association of different SNP genotypes with body weight in Muscovy duck
| SNPs | Genotype | Body weight (kg) | Restriction enzyme | Location |
|---|---|---|---|---|
| g.4819189 C>T | CC (n = 198) | 3.15±0.65 | Hha I | Exon |
| CT (n = 79) | 3.20±0.50 | |||
| TT (n = 57) | 3.05±0.55 | |||
| p-value | p>0.05 | |||
| g.4819220 T>C | TT (n = 239) | 3.05±0.80 | Sac II | Exon |
| TC (n = 68) | 3.10±0.65 | |||
| CC (n = 27) | 2.95±0.50 | |||
| p-value | p>0.05 | |||
| g.4819252 A>G | AA (n = 92) | 2.75c±0.55 | Not I | Intron |
| AG (n = 209) | 3.25a±0.65 | |||
| GG (n = 33) | 3.00b±0.60 | |||
| p-value | p<0.05 | |||
| g.4819303 C>T | CC (n = 150) | 3.05±0.75 | Nco I | Intron |
| CT (n = 38) | 3.10±0.60 | |||
| TT (n = 146) | 3.15±0.65 | |||
| p-value | p>0.05 | |||
| g.4821390 G>A | GG (n = 37) | 2.75B±0.65 | Hha I | Intron |
| GA (n = 235) | 3.15A±0.50 | |||
| AA (n = 62) | 2.70B±0.65 | |||
| p-value | p<0.01 | |||
| g.4825015 C>T | CC (n = 128) | 2.95±0.55 | Sma I | Intron |
| CT (n = 124) | 3.10±0.65 | |||
| TT (n = 85) | 3.05±0.55 | |||
| P-value | p>0.05 | |||
| g.4830197 T>C | TT (n = 234) | 3.05±0.55 | Hha I | 3′UTR |
| TC (n = 54) | 2.90±0.60 | |||
| CC (n = 46) | 2.85±0.55 | |||
| p-value | p>0.05 | |||
| g.4830622 T>G | TT (n = 50) | 3.25a±0.75 | Ban I | 3′UTR |
| TG (n = 63) | 3.00b±0.60 | |||
| GG (n = 221) | 2.85b±0.65 | |||
| p-value | p<0.05 |
REFERENCES
1. Mitchell PO, Mills ST, Pavlath GK. Calcineurin differentially regulates maintenance and growth of phenotypically distinct muscles. Am J Physiol Cell Physiol 2002; 282:C984–92.
https://doi.org/10.1152/ajpcell.00483.2001


2. Iizuka K, Machida T, Hirafuji M. Skeletal muscle is an endocrine organ. J Pharmacol Sci 2014; 125:125–31.
https://doi.org/10.1254/jphs.14r02cp


3. Norring M, Valros A, Valaja J, Sihvo HK, Immonen K, Puolanne E. Wooden breast myopathy links with poorer gait in broiler chickens. Animal 2019; 13:1690–5.
https://doi.org/10.1017/S1751731118003270

4. Glass DJ. Signalling pathways that mediate skeletal muscle hypertrophy and atrophy. Nat Cell Biol 2003; 5:87–90.
https://doi.org/10.1038/ncb0203-87


5. Stewart CE, Rittweger J. Adaptive processes in skeletal muscle: molecular regulators and genetic influences. J Musculoskelet Neuronal Interact 2006; 6:73–86.

6. Grand FL, Rudnicki MA. Skeletal muscle satellite cells and adult myogenesis. Curr Opin Cell Biol 2007; 19:628–33.
https://doi.org/10.1016/j.ceb.2007.09.012



7. Gaali S, Gopalakrishnan R, Wang Y, Kozany C, Hausch F. The chemical biology of immunophilin ligands. Curr Med Chem 2011; 18:5355–79.
https://doi.org/10.2174/092986711798194342


8. Menke A, Klengel T, Rubel J, et al. Genetic variation in FKBP5 associated with the extent of stress hormone dysregulation in major depression. Genes Brain Behav 2013; 12:289–96.
https://doi.org/10.1111/gbb.12026


9. Wang Q, Shelton RC, Dwivedi Y. Interaction between early-life stress and FKBP5 gene variants in major depressive disorder and post-traumatic stress disorder: a systematic review and meta-analysis. J Affect Disord 2018; 225:422–8.
https://doi.org/10.1016/j.jad.2017.08.066


10. O‘Leary JC, Zhang B, Koren J, Blair L, Dickey CA. The role of FKBP5 in mood disorders: Action of FKBP5 on steroid hormone receptors leads to questions about its evolutionary importance. CNS Neurol Disord Drug Targets 2013; 12:1157–62.
https://doi.org/10.2174/187152731131200121



11. Herman JP, McKlveen JM, Ghosal S, et al. Regulation of the hypothalamic–pituitary–adrenocortical stress response. Compr Physiol 2016; 6:603–21.
https://doi.org/10.1002/cphy.c150015



12. Touma C, Gassen NC, Herrmann L, et al. FK506 binding protein 5 shapes stress responsiveness: modulation of neuroendocrine reactivity and coping behavior. Biol Psychiatry 2011; 70:928–36.
https://doi.org/10.1016/j.biopsych.2011.07.023


13. Hartmann J, Wagner KV, Liebl C, et al. The involvement of FK506-binding protein 51 (FKBP5) in the behavioral and neuroendocrine effects of chronic social defeat stress. Neuropharmacology 2012; 62:332–9.
https://doi.org/10.1016/j.neuropharm.2011.07.041


14. Oakley RH, Cidlowski JA. The biology of the glucocorticoid receptor: new signaling mechanisms in health and disease. J Allergy Clin Immunol 2013; 132:1033–44.
https://doi.org/10.1016/j.jaci.2013.09.007



15. Pei HD, Li L, Fridley BL, et al. FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell 2009; 16:259–66.
https://doi.org/10.1016/j.ccr.2009.07.016



16. Gassen NC, Hartmann J, Zschocke J, et al. Association of FKBP51 with priming of autophagy pathways and mediation of antidepressant treatment response: evidence in cells, mice, and humans. PLoS Med 2014; 11:e1001755
https://doi.org/10.1371/journal.pmed.1001755



17. Su AI, Wiltshire T, Batalov S, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA 2004; 101:6062–7.
https://doi.org/10.1073/pnas.0400782101



18. Pereira MJ, Palming J, Svensson MK, et al. FKBP5 expression in human adipose tissue increases following dexamethasone exposure and is associated with insulin resistance. Metabolism 2014; 63:1198–208.
https://doi.org/10.1016/j.metabol.2014.05.015


19. Balsevich G, Uribe A, Wagner KV, et al. Interplay between diet-induced obesity and chronic stress in mice: potential role of FKBP51. J Endocrinol 2014; 222:15–26.
https://doi.org/10.1530/JOE-14-0129


20. Stechschulte LA, Bin Q, Warrier M, et al. FKBP51 null mice are resistant to diet-induced obesity and the PPARgamma agonist rosiglitazone. Endocrinology 2016; 157:3888–900.
https://doi.org/10.1210/en.2015-1996



21. Zannas AS, Balsevich G, Gassen NC. The emerging role of FKBP5 in the regulation of metabolism and body weight. Surg Obes Relat Dis 2016; 12:1560–1.
https://doi.org/10.1016/j.soard.2016.05.016


22. Lu BR, Jiao YL, Wang YC, et al. A FKBP5 mutation is associated with Paget’s disease of bone and enhances osteoclastogenesis. Exp Mol Med 2017; 49:e336
https://doi.org/10.1038/emm.2017.64



23. Hartmann IB, Fries GR, Bucker J, Scotton E, von Diemen L, Kauer-Sant’Anna M. The FKBP5 polymorphism rs1360780 is associated with lower weight loss after bariatric surgery: 26 months of follow-up. Surg Obes Relat Dis 2016; 12:1554–60.
https://doi.org/10.1016/j.soard.2016.04.016


24. Yeo S, Enoch MA, Gorodetsky E, et al. The influence of FKBP5 genotype on expression of FKBP5 and other glucocorticoid-regulated genes, dependent on trauma exposure. Genes Brain Behav 2017; 16:223–32.
https://doi.org/10.1111/gbb.12342


25. Wang G, Zhang S, Wei S, et al. Novel polymorphisms of SIX4 gene and their association with body measurement traits in Qinchuan cattle. Gene 2014; 539:107–10.
https://doi.org/10.1016/j.gene.2014.01.042


26. Zhang C, Chen KK, Zhao XH, Wang C, Geng ZY. Effect of L-theanine on the growth performance, immune function, and jejunum morphology and antioxidant status of ducks. Animal 2019; 13:1145–53.
https://doi.org/10.1017/S1751731118002884


27. Dou TF, Li ZT, Wang K, et al. Regulation of myostatin expression is associated with growth and muscle development in commercial broiler and DMC muscle. Mol Biol Rep 2018; 45:511–22.
https://doi.org/10.1007/s11033-018-4187-7



28. Li H, Zhu C, Tao Z, et al. MyoD and Myf6 gene expression patterns in skeletal muscle during embryonic and posthatch development in the domestic duck (Anas platyrhynchos domestica). J Anim Breed Genet 2014; 131:194–201.
https://doi.org/10.1111/jbg.12057


29. Shimoide T, Kawao N, Tamura Y, Morita H, Kaji H. Novel roles of FKBP5 in muscle alteration induced by gravity change in mice. Biochem Biophys Res Commun 2016; 479:602–6.
https://doi.org/10.1016/j.bbrc.2016.09.126


30. MacMillan D. FK506 binding proteins: cellular regulators of intracellular Ca2+ signalling. Eur J Pharmacol 2013; 700:181–93.
https://doi.org/10.1016/j.ejphar.2012.12.029


31. Garabedian MJ, Harris CA, Jeanneteau F. Glucocorticoid receptor action in metabolic and neuronal function. F1000Res 2017; 6:1208
https://doi.org/10.12688/f1000research.11375.1



32. Laine AL, Laclie C, Furlong J, Crowe MA, Monniaux D. A bovine-specific FSH enzyme immunoassay and its application to study the role of FSH in ovarian follicle development during the postnatal period. Animal 2019; 13:1666–75.
https://doi.org/10.1017/S1751731118003233

33. Shi L, Liu L, Ma Z, et al. Identification of genetic associations of ECHS1 gene with milk fatty acid traits in dairy cattle. Anim Genet 2019; 50:430–8.
https://doi.org/10.1111/age.12833


34. Cao F, Zhang HB, Feng JL, Gao CG, Li SB. Association study of three microsatellite polymorphisms located in introns 1, 8, and 9 of DISC1 with schizophrenia in the Chinese Han population. Genet Test Mol Biomarkers 2013; 17:407–11.
https://doi.org/10.1089/gtmb.2012.0438


35. Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem Sci 2003; 28:182–8.
https://doi.org/10.1016/S0968-0004(03)00051-3


36. Ju ZH, Wang CF, Wang XG, et al. The effect of the SNP g.18475 A>G in the 3′UTR of NCF4 on mastitis susceptibility in dairy cattle. Cell Stress Chaperones 2018; 23:385–91.
https://doi.org/10.1007/s12192-017-0848-z



- TOOLS






 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Supplement
Supplement Print
Print





